Deep generative AI models analyzing circulating orphan non-coding RNAs enable detection of early-stage lung cancer
- PMID: 39572521
- PMCID: PMC11582319
- DOI: 10.1038/s41467-024-53851-9
Deep generative AI models analyzing circulating orphan non-coding RNAs enable detection of early-stage lung cancer
Abstract
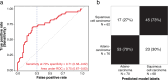
Liquid biopsies have the potential to revolutionize cancer care through non-invasive early detection of tumors. Developing a robust liquid biopsy test requires collecting high-dimensional data from a large number of blood samples across heterogeneous groups of patients. We propose that the generative capability of variational auto-encoders enables learning a robust and generalizable signature of blood-based biomarkers. In this study, we analyze orphan non-coding RNAs (oncRNAs) from serum samples of 1050 individuals diagnosed with non-small cell lung cancer (NSCLC) at various stages, as well as sex-, age-, and BMI-matched controls. We demonstrate that our multi-task generative AI model, Orion, surpasses commonly used methods in both overall performance and generalizability to held-out datasets. Orion achieves an overall sensitivity of 94% (95% CI: 87%-98%) at 87% (95% CI: 81%-93%) specificity for cancer detection across all stages, outperforming the sensitivity of other methods on held-out validation datasets by more than ~ 30%.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors are either employees, shareholders, or stock option holders of Exai Bio, Inc. B.A., H.G., F.H., and M.K. have a pending patent application (U.S. Patent “Systems and Methods for Early-Stage Cancer Detection and Subtyping” Application Serial No. 18/636,128 and International Application No. PCT/US24/24682) related to this work.
Figures



References
-
- American Cancer Society. Lung cancer statistics. https://www.cancer.org/cancer/types/lung-cancer/about/key-statistics.html (2023). Accessed: 2023-01-04.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

