An evolutionarily conserved metabolite inhibits biofilm formation in Escherichia coli K-12
- PMID: 39572562
- PMCID: PMC11582573
- DOI: 10.1038/s41467-024-54501-w
An evolutionarily conserved metabolite inhibits biofilm formation in Escherichia coli K-12
Abstract
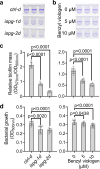
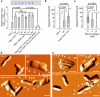
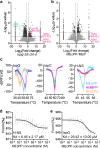
Methylerythritol cyclodiphosphate (MEcPP) is an intermediate in the biosynthesis of isoprenoids in plant plastids and in bacteria, and acts as a stress signal in plants. Here, we show that MEcPP regulates biofilm formation in Escherichia coli K-12 MG1655. Increased MEcPP levels, triggered by genetic manipulation or oxidative stress, inhibit biofilm development and production of fimbriae. Deletion of fimE, encoding a protein known to downregulate production of adhesive fimbriae, restores biofilm formation in cells with elevated MEcPP levels. Limited proteolysis-coupled mass spectrometry (LiP-MS) reveals that MEcPP interacts with the global regulatory protein H-NS, which is known to repress transcription of fimE. MEcPP prevents the binding of H-NS to the fimE promoter. Therefore, our results indicate that MEcPP can regulate biofilm formation by modulating H-NS activity and thus reducing fimbriae production. Further research is needed to test whether MEcPP plays similar regulatory roles in other bacteria.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli.Microbiology (Reading). 2005 Oct;151(Pt 10):3287-3298. doi: 10.1099/mic.0.28098-0. Microbiology (Reading). 2005. PMID: 16207912
-
Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12.Microbiology (Reading). 2020 Sep;166(9):880-890. doi: 10.1099/mic.0.000947. Microbiology (Reading). 2020. PMID: 32649279
-
Disruption of rcsB by a duplicated sequence in a curli-producing Escherichia coli O157:H7 results in differential gene expression in relation to biofilm formation, stress responses and metabolism.BMC Microbiol. 2017 Mar 8;17(1):56. doi: 10.1186/s12866-017-0966-x. BMC Microbiol. 2017. PMID: 28274217 Free PMC article.
-
Plastidial metabolites and retrograde signaling: A case study of MEP pathway intermediate MEcPP that orchestrates plant growth and stress responses.Plant Physiol Biochem. 2025 May;222:109747. doi: 10.1016/j.plaphy.2025.109747. Epub 2025 Mar 5. Plant Physiol Biochem. 2025. PMID: 40073740 Review.
-
Contribution of rpoS and bolA genes in biofilm formation in Escherichia coli K-12 MG1655.Mol Cell Biochem. 2010 Sep;342(1-2):207-13. doi: 10.1007/s11010-010-0485-7. Epub 2010 May 18. Mol Cell Biochem. 2010. PMID: 20480211 Review.
Cited by
-
Mapping protein-metabolite interactions in E. coli by integrating chromatographic techniques and co-fractionation mass spectrometry.iScience. 2025 May 8;28(6):112611. doi: 10.1016/j.isci.2025.112611. eCollection 2025 Jun 20. iScience. 2025. PMID: 40491478 Free PMC article.
References
-
- Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol.56, 187–209 (2002). - PubMed
-
- Teschler, J. K., Nadell, C. D., Drescher, K. & Yildiz, F. H. Mechanisms underlying vibrio cholerae biofilm formation and dispersion. Annu. Rev. Microbiol.76, 503–532 (2022). - PubMed
-
- Jenal, U., Reinders, A. & Lori, C. Cyclic di-GMP: second messenger extraordinaire. Nat. Rev. Microbiol.15, 271–284 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

