Acid-exposed and hypoxic cancer cells do not overlap but are interdependent for unsaturated fatty acid resources
- PMID: 39572570
- PMCID: PMC11582563
- DOI: 10.1038/s41467-024-54435-3
Acid-exposed and hypoxic cancer cells do not overlap but are interdependent for unsaturated fatty acid resources
Abstract
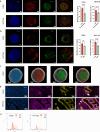
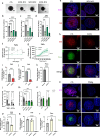
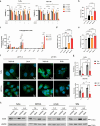
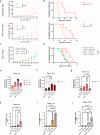
Cancer cells in acidic tumor regions are aggressive and a key therapeutic target, but distinguishing between acid-exposed and hypoxic cells is challenging. Here, we use carbonic anhydrase 9 (CA9) antibodies to mark acidic areas in both hypoxic and respiring tumor areas, along with an HRE-GFP reporter for hypoxia, to isolate distinct cell populations from 3D tumor spheroids. Transcriptomic analysis of CA9-positive, hypoxia-negative cells highlights enriched fatty acid desaturase activity. Inhibiting or silencing stearoyl-CoA desaturase-1 (SCD1) induces ferroptosis in CA9-positive acidic cancer cells and delays mouse tumor growth, an effect enhanced by omega-3 fatty acid supplementation. Using acid-exposed cancer cells and patient-derived tumor organoids, we show that SCD1 inhibition increases acidic cancer cell reliance on external mono-unsaturated fatty acids, depriving hypoxic cells of essential resources. This bystander effect provides unbiased evidence for a lack of full overlap between hypoxic and acidic tumor compartments, highlighting a rationale for targeting desaturase activity in cancer.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Corbet, C. & Feron, O. Tumour acidosis: from the passenger to the driver’s seat. Nat. Rev. Cancer17, 577–593 (2017). - PubMed
-
- Swietach, P., Boedtkjer, E. & Pedersen, S. F. How protons pave the way to aggressive cancers. Nat. Rev. Cancer23, 825–841 (2023). - PubMed
-
- Corbet, C. & Feron, O. Cancer cell metabolism and mitochondria: nutrient plasticity for TCA cycle fueling. Biochim. Biophys. Acta Rev. Cancer1868, 7–15 (2017). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

