Personalised antimicrobial susceptibility testing with clinical prediction modelling informs appropriate antibiotic use
- PMID: 39572574
- PMCID: PMC11582675
- DOI: 10.1038/s41467-024-54192-3
Personalised antimicrobial susceptibility testing with clinical prediction modelling informs appropriate antibiotic use
Abstract
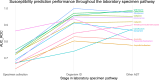
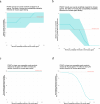
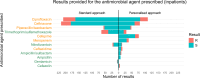
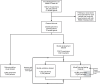
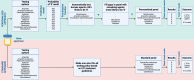
Antimicrobial susceptibility testing is a key weapon against antimicrobial resistance. Diagnostic microbiology laboratories use one-size-fits-all testing approaches that are often imprecise, inefficient, and inequitable. Here, we report a personalised approach that adapts laboratory testing for urinary tract infection to maximise the number of appropriate treatment options for each patient. We develop and assess susceptibility prediction models for 12 antibiotics on real-world healthcare data using an individual-level simulation study. When combined with decision thresholds that prioritise selection of World Health Organisation Access category antibiotics (those least likely to induce antimicrobial resistance), the personalised approach delivers more susceptible results (results that encourage prescription of that antibiotic) per specimen for Access category antibiotics than a standard testing approach, without compromising provision of susceptible results overall. Here, we show that personalised antimicrobial susceptibility testing could help tackle antimicrobial resistance by safely providing more Access category antibiotic treatment options to clinicians managing urinary tract infection.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: Alex Howard declares personal consulting work for Pfizer outside the submitted work, and a donation from Pfizer to the University of Liverpool for a public and professional engagement project outside the submitted work. Iain Buchan declares consulting fees via University of Liverpool from AstraZeneca outside the submitted work. William Hope holds or has recently held research grants with UKRI, EU (FP7, IMI-1, IMI-2), Wellcome, F2G, Spero Therapeutics, Antabio, Pfizer, Allecra, Bugworks, Phico Therapeutics, BioVersys, Global Antimicrobial Research and Development Partnership (GARDP). He is (or has recently been) a consultant for Appili Therapeutics, F2G, Spero Therapeutics, Pfizer, GSK, Phico Therapeutics, Pulmocide, and Mundipharma Research Ltd. He was a member of the Specialist Advisory Committee for GARDP (2020-2023), a member of the British Society for Antimicrobial Chemotherapy (BSAC) Breakpoint Committee (2020-2023), a member of Health Technology Appraisal (HTA) Prioritisation Committee for hospital care and was the Specialty National Co-lead for Infection for the National Institute of Health Research (NIHR) (2020-2024). The other authors declare no competing interests.
Figures



References
-
- United Nations General Assembly. Political Declaration of the High-level Meeting on Antimicrobial Resistance, 9 September 2024.https://www.un.org/pga/wp-content/uploads/sites/108/2024/09/FINAL-Text-A... (2024).
-
- WHO Control and Response Strategies Team, WHO Surveillance, Prevention and Control Team. Global Research Agenda for Antimicrobial Resistance in Human Health.https://www.who.int/publications/m/item/global-research-agenda-for-antim... (2023).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

