GPATCH11 variants cause mis-splicing and early-onset retinal dystrophy with neurological impairment
- PMID: 39572588
- PMCID: PMC11582697
- DOI: 10.1038/s41467-024-54549-8
GPATCH11 variants cause mis-splicing and early-onset retinal dystrophy with neurological impairment
Abstract
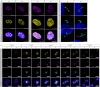
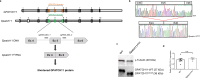
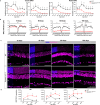
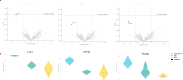
Here we conduct a study involving 12 individuals with retinal dystrophy, neurological impairment, and skeletal abnormalities, with special focus on GPATCH11, a lesser-known G-patch domain-containing protein, regulator of RNA metabolism. To elucidate its role, we study fibroblasts from unaffected individuals and patients carrying the recurring c.328+1 G > T mutation, which specifically removes the main part of the G-patch domain while preserving the other domains. Additionally, we generate a mouse model replicating the patients' phenotypic defects, including retinal dystrophy and behavioral abnormalities. Our results reveal a subcellular localization of GPATCH11 characterized by a diffuse presence in the nucleoplasm, as well as centrosomal localization, suggesting potential functions in RNA and cilia metabolism. Transcriptomic analysis performed on mouse retina detect dysregulation in both gene expression and splicing activity, impacting key processes such as photoreceptor light responses, RNA regulation, and primary cilia-associated metabolism. Proteomic analysis of mouse retina confirms the roles GPATCH11 plays in RNA processing, splicing, and transcription regulation, while also suggesting additional functions in synaptic plasticity and nuclear stress response. Our research provides insights into the diverse roles of GPATCH11 and identifies that the mutations affecting this protein are responsible for a recently characterized described syndrome.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Bohnsack, K. E., Ficner, R., Bohnsack, M. T. & Jonas, S. Regulation of DEAH-box RNA helicases by G-patch proteins. Biol. Chem.402, 561–579 (2021). - PubMed
-
- Abdelhaleem, M. Helicases: an Overview. in Helicases (ed. Abdelhaleem, M. M.) 587 1–12 (Humana Press, 2009).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

