The shared genetic architecture and evolution of human language and musical rhythm
- PMID: 39572686
- PMCID: PMC11860242
- DOI: 10.1038/s41562-024-02051-y
The shared genetic architecture and evolution of human language and musical rhythm
Abstract
This study aimed to test theoretical predictions over biological underpinnings of previously documented phenotypic correlations between human language-related and musical rhythm traits. Here, after identifying significant genetic correlations between rhythm, dyslexia and various language-related traits, we adapted multivariate methods to capture genetic signals common to genome-wide association studies of rhythm (N = 606,825) and dyslexia (N = 1,138,870). The results revealed 16 pleiotropic loci (P < 5 × 10-8) jointly associated with rhythm impairment and dyslexia, and intricate shared genetic and neurobiological architectures. The joint genetic signal was enriched for foetal and adult brain cell-specific regulatory regions, highlighting complex cellular composition in their shared underpinnings. Local genetic correlation with a key white matter tract (the left superior longitudinal fasciculus-I) substantiated hypotheses about auditory-motor connectivity as a genetically influenced, evolutionarily relevant neural endophenotype common to rhythm and language processing. Overall, we provide empirical evidence of multiple aspects of shared biology linking language and musical rhythm, contributing novel insight into the evolutionary relationships between human musicality and linguistic communication traits.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: P.F. is employed by and holds stock or stock options in 23andMe, Inc. The other authors declare no competing interests.
Figures
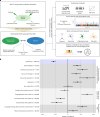
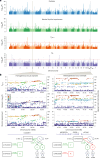
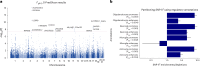

Update of
-
The shared genetic architecture and evolution of human language and musical rhythm.bioRxiv [Preprint]. 2023 Nov 2:2023.11.01.564908. doi: 10.1101/2023.11.01.564908. bioRxiv. 2023. Update in: Nat Hum Behav. 2025 Feb;9(2):376-390. doi: 10.1038/s41562-024-02051-y. PMID: 37961248 Free PMC article. Updated. Preprint.
References
-
- Tarar, J. M., Meisinger, E. B. & Dickens, R. H. Test review: test of word reading efficiency–second edition (TOWRE-2) by Torgesen, J. K., Wagner, R. K. & Rashotte, C. A. Can. J. Sch. Psychol.30, 320–326 (2015). - DOI
MeSH terms
Grants and funding
- R01 DC016977/DC/NIDCD NIH HHS/United States
- DP2 HD098859/HD/NICHD NIH HHS/United States
- K18 DC017383/DC/NIDCD NIH HHS/United States
- P50 HD103537/HD/NICHD NIH HHS/United States
- R01DC016977/U.S. Department of Health & Human Services | NIH | National Institute on Deafness and Other Communication Disorders (NIDCD)
LinkOut - more resources
Full Text Sources

