Integrative proteomic analyses across common cardiac diseases yield mechanistic insights and enhanced prediction
- PMID: 39572695
- PMCID: PMC11634769
- DOI: 10.1038/s44161-024-00567-0
Integrative proteomic analyses across common cardiac diseases yield mechanistic insights and enhanced prediction
Abstract
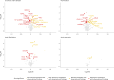
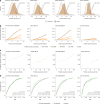
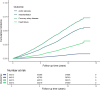
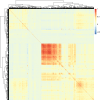
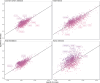
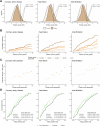
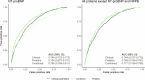
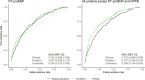
Cardiac diseases represent common highly morbid conditions for which molecular mechanisms remain incompletely understood. Here we report the analysis of 1,459 protein measurements in 44,313 UK Biobank participants to characterize the circulating proteome associated with incident coronary artery disease, heart failure, atrial fibrillation and aortic stenosis. Multivariable-adjusted Cox regression identified 820 protein-disease associations-including 441 proteins-at Bonferroni-adjusted P < 8.6 × 10-6. Cis-Mendelian randomization suggested causal roles aligning with epidemiological findings for 4% of proteins identified in primary analyses, prioritizing therapeutic targets across cardiac diseases (for example, spondin-1 for atrial fibrillation and the Kunitz-type protease inhibitor 1 for coronary artery disease). Interaction analyses identified seven protein-disease associations that differed Bonferroni-significantly by sex. Models incorporating proteomic data (versus clinical risk factors alone) improved prediction for coronary artery disease, heart failure and atrial fibrillation. These results lay a foundation for future investigations to uncover disease mechanisms and assess the utility of protein-based prevention strategies for cardiac diseases.
© 2024. The Author(s).
Conflict of interest statement
Inclusion and ethics: Inclusion and ethics standards have been reviewed where applicable. Competing interests: J.L.J. reports board membership of Imbria Pharmaceuticals; grant support from Abbott Diagnostics, AstraZeneca, BMS, HeartFlow and Novartis; previous consulting income from Abbott Diagnostics, AstraZeneca, Bayer, Beckman Coulter, Jana Care, Janssen, Novartis, Quidel, Roche Diagnostics and Siemens; and clinical end point committee/data safety monitoring board membership for Abbott, Bayer, AbbVie, CVRx, Pfizer, Roche Diagnostics and Takeda. M.C.H. reports consulting fees from CRISPR Therapeutics and Comanche Biopharma; advisory board service for Miga Health; and grant support from Genentech. P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech/Roche and Novartis; personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Esperion Therapeutics, Foresite Capital, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, Novartis, TenSixteen Bio and Tourmaline Bio; equity in Bolt, Candela, Mercury, MyOme, Parameter Health, Preciseli and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. The other authors declare no competing interests.
Figures






Update of
-
Integrative proteomic analyses across common cardiac diseases yield new mechanistic insights and enhanced prediction.medRxiv [Preprint]. 2023 Dec 19:2023.12.19.23300218. doi: 10.1101/2023.12.19.23300218. medRxiv. 2023. Update in: Nat Cardiovasc Res. 2024 Dec;3(12):1516-1530. doi: 10.1038/s44161-024-00567-0. PMID: 38196601 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- K08 HL166687/HL/NHLBI NIH HHS/United States
- R01 HL127564/HL/NHLBI NIH HHS/United States
- R01HL127564/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- 979465/American Heart Association (American Heart Association, Inc.)
- R00 HG012956/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
