The gut microbiota of three avian species living in sympatry
- PMID: 39574002
- PMCID: PMC11580620
- DOI: 10.1186/s12862-024-02329-9
The gut microbiota of three avian species living in sympatry
Abstract
Background: Evolutionary divergence and genetic variation are often linked to differences in microbial community structure and diversity. While environmental factors and diet heavily influence gut microbial communities, host species contributions are harder to quantify. Closely related species living in sympatry provide a unique opportunity to investigate species differences without the confounding effects of habitat and dietary variation. We therefore compared and contrasted the gut microbiota of three sympatric plover species: the widespread Kittlitz's and white-fronted plovers (Anarhynchus pecuarius and A. marginatus) and the endemic and vulnerable Madagascar plover (A. thoracicus).
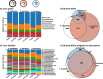
Results: We found no significant differences in the beta diversity (composition) of the gut microbiota of the three species. However, A. thoracicus exhibited higher intraspecific compositional similarity (i.e. lower pairwise distances) than the other two species; this pattern was especially pronounced among juveniles. By contrast, microbial alpha diversity varied significantly among the species, being highest in A. pecuarius, intermediate in A. marginatus and lowest in A. thoracicus. This pattern was again stronger among juveniles. Geographical distance did not significantly affect the composition of the gut microbiota, but genetic relatedness did.
Conclusion: While patterns of microbial diversity varied across species, the lack of compositional differences suggests that habitat and diet likely exert a strong influence on the gut microbiota of plovers. This may be enhanced by their precocial, ground-dwelling nature, which could facilitate the horizontal transmission of microbes from the environment. We hypothesise that gut microbiota diversity in plovers primarily reflects the ecological pool of microbiota, which is subsequently modified by host-specific factors including genetics. The reduced microbial and genetic diversity of the endemic A. thoracicus may hinder its ability to adapt to environmental changes, highlighting the need for increased conservation efforts for this vulnerable species.
Keywords: Endemic; Genetics; Gut microbiota; Holobiont; Madagascar; Plovers.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Sampling of the three species of Madagascar plovers was approved by the Direction des Aires Protégées, des Ressources Naturellement Renouvelables et des Ecosystèmes from Madagascar (permits n $$^{\circ }$$ ∘ 386/21 and 054/21/MEDD/SG/DGGE/DAPRNE/SCBE.Re). Consent for publication: Not applicable. Completing interests: The authors declare no competing interests.
Figures




References
-
- Rosenberg E. Microbiomes: Current Knowledge and Unanswered Questions. Cham: Springer Nature Switzerland AG; 2021.
-
- Grond K, Sandercock BK, Jumpponen A, Zeglin LH. The avian gut microbiota: community, physiology and function in wild birds. J Avian Biol. 2018;49(11): e01788. - DOI
MeSH terms
Grants and funding
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 316099922/Deutsche Forschungsgemeinschaft
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
- 396780709/Deutsche Forschungsgemeinschaft,
LinkOut - more resources
Full Text Sources
