Investigation the role of SIRT3, SIRT7, NFATC1, and PDL-1 genes in androgenetic alopecia
- PMID: 39574094
- PMCID: PMC11583656
- DOI: 10.1186/s13104-024-06980-9
Investigation the role of SIRT3, SIRT7, NFATC1, and PDL-1 genes in androgenetic alopecia
Abstract
Background: Androgenetic alopecia (AGA) stands as the most prevalent form of hair loss, affecting the hair follicles (HFs). Aging emerges as a prominent contributor in this condition. In this study, our aim is to elucidate the expression patterns of candidate genes-SIRT3, SIRT7, NFATC1, and PDL-1-known to exhibit differential expression levels during HF aging, and to underscore the role of aging in AGA.
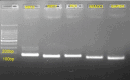
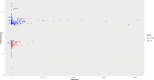
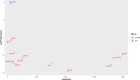
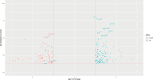
Material and methods: Mesenchymal stem cells (MSCs) were isolated from the vertex and occipital regions of six men affected by AGA. The aim was to assess the expression levels of SIRT3, SIRT7, NFATC1, and PDL-1 genes. RNA extraction was performed followed by cDNA synthesis, and gene expression levels were quantified using real-time PCR. To validate the experimental findings, two different RNA-seq datasets relevant to the study were analyzed using R software.
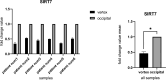
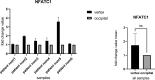
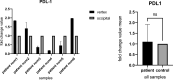
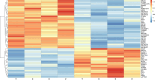
Results: In the present study, experimental tests revealed that the expression levels of SIRT3 and SIRT7, known to decrease during HF aging, were diminished in AGA-affected samples as well. Conversely, the mean value of NFATC1 and PDL-1 expression level, which are known to increase during HF aging, were found to be elevated in AGA-affected samples. Moreover, bioinformatic analyses provide additional support for the role of SIRT3, SIRT7 and NFATC1in AGA pathogenesis.
Conclusion: While SIRT3 and SIRT7 may play critical roles in AGA development, further research is needed to elucidate the significance of NFATC1 and PDL-1 in this context and to explore their potential as therapeutic targets for AGA treatment.
Keywords: NFATC1; PDL1; SIRT3; SIRT7; Androgenetic alopecia.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: This research was approved by the Ethics Committee of Tarbiat Modares University (IR.MODARES.REC.1402.244), Tehran, Iran. All the participants have accepted and signed the informed consent during the standard genetic counselling sessions. Written informed consent was obtained from the patients for publication of this study. A copy of the written consent is available for review by the Editor of this journal. Consent for publication: All authors reviewed the results and approved the final version of the manuscript. Competing interests: The authors declare no competing interests.
Figures



References
-
- Kidangazhiathmana A, Santhosh P. Pathogenesis of androgenetic alopecia. Clin Dermatol Rev. 2022;6(2):69–74. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

