This is a preprint.
Paired and solitary ionocytes in the zebrafish olfactory epithelium
- PMID: 39574570
- PMCID: PMC11580993
- DOI: 10.1101/2024.11.08.620918
Paired and solitary ionocytes in the zebrafish olfactory epithelium
Update in
-
Paired and solitary ionocytes in the zebrafish olfactory epithelium.Chem Senses. 2025 Jan 22;50:bjaf031. doi: 10.1093/chemse/bjaf031. Chem Senses. 2025. PMID: 40810206 Free PMC article.
Abstract
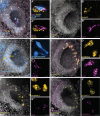
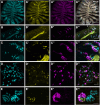
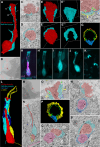
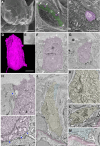
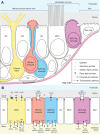
The sense of smell is generated by electrical currents that are influenced by the concentration of ions in olfactory sensory neurons and mucus. In contrast to the extensive morphological and molecular characterization of sensory neurons, there has been little description of the cells that control ion concentrations in the zebrafish olfactory system. Here, we report the molecular and ultrastructural characterization of zebrafish olfactory ionocytes. Transcriptome analysis suggests that the zebrafish olfactory epithelium contains at least three different ionocyte types, which resemble Na+/K+-ATPase-rich (NaR), H+-ATPase-rich (HR), and Na+/Cl- cotransporter (NCC) cells, responsible for calcium, pH, and chloride regulation, respectively, in the zebrafish skin. In the olfactory epithelium, NaR-like and HR-like ionocytes are usually adjacent to one another, whereas NCC-like cells are usually solitary. The distinct subtypes are differentially distributed: NaR-like/HR-like cell pairs are found broadly within the olfactory epithelium, whereas NCC-like cells reside within the peripheral non-sensory multiciliated cell zone. Comparison of gene expression and serial-section electron microscopy analysis indicates that the NaR-like cells wrap around the HR-like cells and are connected to them by shallow tight junctions. The development of olfactory ionocyte subtypes is also differentially regulated, as pharmacological Notch inhibition leads to a loss of NaR-like and HR-like cells, but does not affect NCC-like ionocyte number. These results provide a molecular and anatomical characterization of olfactory ionocytes in a stenohaline freshwater teleost. The paired ionocytes suggest that both transcellular and paracellular transport regulate ion concentrations in the olfactory epithelium, while the solitary ionocytes may enable independent regulation of ciliary beating.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures





References
-
- Bertmar G. 1972. Labyrinth cells, a new cell type in vertebrate olfactory organs. Z Für Zellforsch Mikrosk Anat. 132:245–256. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
