This is a preprint.
PgtE protease enables virulent Salmonella to evade C3-mediated serum and neutrophil killing
- PMID: 39574608
- PMCID: PMC11580845
- DOI: 10.1101/2024.11.05.622138
PgtE protease enables virulent Salmonella to evade C3-mediated serum and neutrophil killing
Update in
-
PgtE protease enables virulent Salmonella to evade C3-mediated serum and neutrophil killing.mBio. 2025 Aug 13;16(8):e0380224. doi: 10.1128/mbio.03802-24. Epub 2025 Jul 14. mBio. 2025. PMID: 40767554 Free PMC article.
Abstract
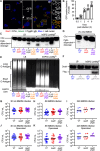
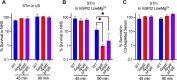
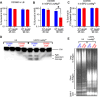
Non-typhoidal Salmonella serovars, such as Salmonella enterica serovar Typhimurium (STm), are a leading cause of inflammatory diarrhea in otherwise healthy individuals. Among children, the elderly, and immunocompromised individuals, STm can spread to systemic sites and cause potentially lethal bacteremia. Phagocytic cells and the immune complement system are pivotal to preventing the dissemination of STm. PgtE, an STm outer membrane protease, has been previously described to cleave over a dozen mammalian protein substrates in vitro, including complement protein C3. However, these activities have mostly been observed with mutant, avirulent strains with a truncated O-antigen that renders bacteria sensitive to complement killing. Here, we report that virulent STm utilizes PgtE to evade complement-mediated killing in vivo. The wild-type pathogen increases pgtE expression and PgtE proteolytic function within macrophages and in macrophage-like in vitro growth conditions, concomitant with physiologic O-antigen shortening in these environments. Furthermore, we found that wild-type STm's resistance to complement-mediated serum and neutrophil killing is PgtE-dependent. We propose that PgtE promotes the systemic spread of STm by acting as a second line of defense against complement when STm escapes from a macrophage.
Figures


References
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. - PubMed
-
- Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin Infect Dis 32:263–269. - PubMed
-
- Lester A, Eriksen NH, Nielsen H, Nielsen PB, Friis-Møller A, Bruun B, Scheibel J, Gaarslev K, Kolmos HJ. 1991. Non-typhoid Salmonella bacteraemia in Greater Copenhagen 1984 to 1988. Eur J Clin Microbiol Infect Dis 10:486–490. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
