This is a preprint.
Systematic review and meta-analysis of bulk RNAseq studies in human Alzheimer's disease brain tissue
- PMID: 39574617
- PMCID: PMC11580990
- DOI: 10.1101/2024.11.07.622520
Systematic review and meta-analysis of bulk RNAseq studies in human Alzheimer's disease brain tissue
Update in
-
Systematic review and meta-analysis of bulk RNAseq studies in human Alzheimer's disease brain tissue.Alzheimers Dement. 2025 Mar;21(3):e70025. doi: 10.1002/alz.70025. Alzheimers Dement. 2025. PMID: 40042520 Free PMC article.
Abstract
Objective: To systematically review and meta-analyze bulk RNA sequencing studies comparing Alzheimer's disease (AD) patients with controls in human brain tissue, assessing study quality and identifying key genes and pathways.
Methods: We searched PubMed, Web of Science, and Scopus on September 23, 2023, for studies using bulk RNAseq on primary human brain tissue from AD patients and controls. Excluded were non-primary tissue, re-analyses without new data, limited RNA types and gene panels. Quality was assessed with a 10-category tool. Meta-analysis used high-quality datasets.
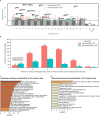
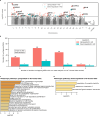
Results: From 3,266 records, 24 studies met criteria. Meta-analysis found 571 differentially expressed genes (DEGs) in temporal lobe and 189 in frontal lobe; overlapping pathways included "Tube morphogenesis" and "Neuroactive ligand-receptor interaction."
Limitations: Study heterogeneity and limited data tables constrained the review.
Conclusions: Rigorous methods are vital in AD transcriptomic studies. Findings enhance understanding of transcriptomic changes, aiding biomarker and therapeutic development.
Registration: PROSPERO (CRD42023466522).
Keywords: Alzheimer’s disease (AD); RNA sequencing (RNAseq); differentially expressed genes (DEGs); human brain tissue; meta-analysis; systematic review.
Conflict of interest statement
Competing interests Statement: The authors report no competing interests.
Figures

References
-
- 2023. Alzheimer’s disease facts and figures - 2023 - Alzheimer’s & Dementia - Wiley Online Library. https://alz-journals.onlinelibrary.wiley.com/doi/10.1002/alz.13016. - DOI - PubMed
-
- Dementia. https://www.who.int/news-room/fact-sheets/detail/dementia.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
