This is a preprint.
Constitutive activity of an atypical chemokine receptor revealed by inverse agonistic nanobodies
- PMID: 39574661
- PMCID: PMC11580867
- DOI: 10.1101/2024.11.04.621790
Constitutive activity of an atypical chemokine receptor revealed by inverse agonistic nanobodies
Abstract
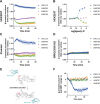
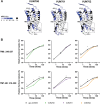
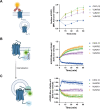
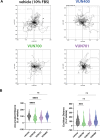
Chemokine stimulation of atypical chemokine receptor 3 (ACKR3) does not activate G proteins but recruits arrestins. It is a chemokine scavenger that indirectly influences responses by restricting the availability of CXCL12, an agonist shared with the canonical receptor CXCR4. ACKR3 is upregulated in numerous disorders. Due to limited insights in chemokine-activated ACKR3 signaling, it is unclear how ACKR3 contributes to pathological phenotypes. One explanation may be that high constitutive activity of ACKR3 drives non-canonical signaling through a basal receptor state. Here we characterize the constitutive action of ACKR3 using novel inverse agonistic nanobodies to suppress basal activity. These new tools promote an inactive receptor conformation which decreased arrestin engagement and inhibited constitutive internalization. Basal, non-chemotactic, breast cancer cell motility was also suppressed, suggesting a role for ACKR3 in this process. The basal receptor activity in pathophysiology may provide a new therapeutic approach for targeting ACKR3.
Conflict of interest statement
Competing interests B.F.V. has an ownership interest in Protein Foundry, L.L.C. and XLock Biosciences, Inc. R.H. is affiliated with QVQ Holding BV. All other authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
