This is a preprint.
TIA1 Mediates Divergent Inflammatory Responses to Tauopathy in Microglia and Macrophages
- PMID: 39574689
- PMCID: PMC11580906
- DOI: 10.1101/2024.11.06.622325
TIA1 Mediates Divergent Inflammatory Responses to Tauopathy in Microglia and Macrophages
Abstract
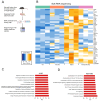
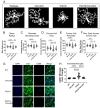
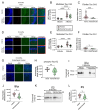
The RNA binding protein TIA1 is known to regulate stress responses. Here we show that TIA1 plays a much broader role in inflammatory cells, being required for the microglial sensome. We crossed TIA1 cKO mice (using a CX3CR1 driven cre element) to PS19 MAPT P301S tauopathy mice. The peripheral macrophages of TIA1 cKO mice exhibited a hyper-inflammatory phenotype with increased cytokine signaling, as expected. Surprisingly, the brains of these mice showed striking reductions in inflammation, including decreases in microglial inflammatory cytokines (TNFα and IL-1β) and sensome markers (CLEC7A, TREM2, ITGAX); these reductions were accompanied by corresponding decreases in tau pathology. Analysis of the brain TIA1 protein interactome identified brain selective TIA1 protein mediated pathways, including strong interactions with the microglial protein C1q, which directs pruning of dystrophic neurons. These results uncover a previously unknown regulatory role for TIA1 in microglial activation in the context of neurodegenerative disease and highlights the divergent regulation of two mononuclear phagocytic lineages: microglia and macrophages.
Conflict of interest statement
Competing interests B.W. is co-founder and Chief Scientific Officer for Aquinnah Pharmaceuticals Inc.
Figures




References
-
- Anderson P., et al. A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J Immunol 144, 574–582 (1990). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
