This is a preprint.
DNA repair and anti-cancer mechanisms in the long-lived bowhead whale
- PMID: 39574710
- PMCID: PMC11580846
- DOI: 10.1101/2023.05.07.539748
DNA repair and anti-cancer mechanisms in the long-lived bowhead whale
Abstract
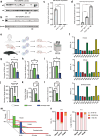
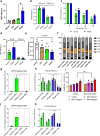
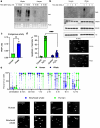
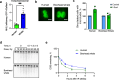
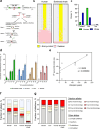
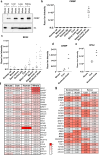
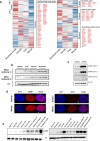
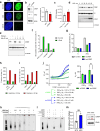
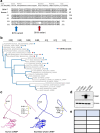
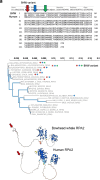
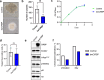
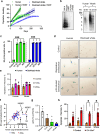
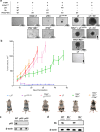
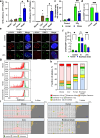
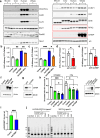
At over 200 years, the maximum lifespan of the bowhead whale exceeds that of all other mammals. The bowhead is also the second-largest animal on Earth, reaching over 80,000 kg1. Despite its very large number of cells and long lifespan, the bowhead is not highly cancer-prone, an incongruity termed Peto's Paradox2. This phenomenon has been explained by the evolution of additional tumor suppressor genes in other larger animals, supported by research on elephants demonstrating expansion of the p53 gene3-5. Here we show that bowhead whale fibroblasts undergo oncogenic transformation after disruption of fewer tumor suppressors than required for human fibroblasts. However, analysis of DNA repair revealed that bowhead cells repair double strand breaks (DSBs) and mismatches with uniquely high efficiency and accuracy compared to other mammals. The protein CIRBP, implicated in protection from genotoxic stress, was present in very high abundance in the bowhead whale relative to other mammals. We show that CIRBP and its downstream protein RPA2, also present at high levels in bowhead cells, increase the efficiency and fidelity of DNA repair in human cells. These results indicate that rather than possessing additional tumor suppressor genes as barriers to oncogenesis, the bowhead whale relies on more accurate and efficient DNA repair to preserve genome integrity. This strategy which does not eliminate damaged cells but repairs them may be critical for the long and cancer-free lifespan of the bowhead whale.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures

References
-
- Peto R. Epidemiology, multistage models, and short-term mutagenicity tests. in The origins of human cancer (eds. Hiatt H., Watson J. & Winsten J.) vol. 4 1403–1428 (Cold Spring Harbor Conferences on Cell Proliferation, NY, 1977).
-
- Vazquez J. M., Sulak M., Chigurupati S. & Lynch V. J. A Zombie LIF Gene in Elephants Is Upregulated by TP53 to Induce Apoptosis in Response to DNA Damage. Cell Rep 24, 1765–1776 (2018). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
