This is a preprint.
Antigen-Specific T Cell Receptor Discovery for Treating Progressive Multifocal Leukoencephalopathy
- PMID: 39574748
- PMCID: PMC11580961
- DOI: 10.1101/2024.11.04.621904
Antigen-Specific T Cell Receptor Discovery for Treating Progressive Multifocal Leukoencephalopathy
Abstract
Background: Progressive multifocal leukoencephalopathy (PML) is a frequently fatal disease of the central nervous system caused by JC virus (JCV). Survival is dependent on early diagnosis and ability to re-establish anti-viral T cell immunity. Adoptive transfer of polyomavirus-specific T cells has shown promise; however, there are no readily available HLA-matched anti-viral T cells to facilitate rapid treatment.
Objective: Identify epitopes of the JCV major capsid protein VP1 that elicit an immune response in the context of human leukocyte antigen allele A*02:01 (HLA-A2) and isolate cognate T cell receptors (TCRs) from healthy donors. Evaluate individual VP1-specific TCRs for their capacity to be expressed in T cells and clear JCV in vitro.
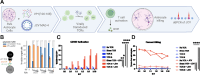
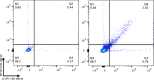
Methods: PBMCs from HLA-A2+ healthy donors were stimulated with peptide libraries tiled across the JCV VP1 protein. Multiple rounds of stimulation were performed to identify the antigens that induced the largest expansion and CD8+ T cell response (measured as INFγ, TNFα, CD137, and CD69 expression). High-affinity, antigen-specific CD8+ T cells were isolated based on intensity of tetramer binding for downstream single-cell TCR sequencing. Candidate TCRs were selected based on tetramer binding affinity and activation assays. Promising TCRs were introduced into the T cell genome via viral transduction for in vitro validation including peptide-pulsed K562 cells and astrocyte cells, and JCV-infected astrocytes.
Results: Four conserved JCV VP1 epitopes (amino acids 100-108, 251-259, 253-262, and 274-283) presented by HLA-A2 were identified. VP1(100-108) consistently elicited the highest level of IFN-γ production from multiple donors and this peptide is in a highly conserved region of VP1. We next identified fourteen high avidity TCRs specific for VP1(100-108). When virally transduced into primary human T cells, seven of these TCRs demonstrated specific binding to VP1(100-108):HLA-A2 tetramers, and four showed increased IFN-γ response when incubated with peptide. Primary CD8+ T cells expressing two of these TCRs cleared both HLA-A2 positive K562 cells and HLA-A2 positive SVG astrocyte cell line presenting exogenously added VP1 peptide at a range of E:T ratios. In addition, both TCR-transduced T cell populations effectively lysed JCV-infected astrocytes.
Conclusions: We identified JCV VP1 epitopes that are immunogenic in the context of HLA-A2 MHC-I, including epitopes that have not been previously described. The VP1(100-108) epitope was used to isolate HLA-A2-restricted TCRs. When cloned into primary human CD8+ T cells, these TCRs recognized VP1 (100-108)-presenting targets, and the transduced T cells conferred cytotoxic activity and eliminated K562 and astrocyte cells displaying the VP1(100-108) peptide and not sham peptide, as well as JCV-infected astrocytes. Taken together, these data suggest that JCV VP1-specific TCRs could be appealing therapeutics for HLA-A2+ individuals with PML in whom intrinsic T cell immunity cannot be rescued.
Figures


References
-
- Padgett B.L., et al. , Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet, 1971. 1(7712): p. 1257–60. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
