Ensemble modeling of SARS-CoV-2 immune dynamics in immunologically naïve rhesus macaques predicts that potent, early innate immune responses drive viral elimination
- PMID: 39575237
- PMCID: PMC11578959
- DOI: 10.3389/fimmu.2024.1426016
Ensemble modeling of SARS-CoV-2 immune dynamics in immunologically naïve rhesus macaques predicts that potent, early innate immune responses drive viral elimination
Abstract
Introduction: An unprecedented breadth of longitudinal viral and multi-scale immunological data has been gathered during SARS-CoV-2 infection. However, due to the high complexity, non-linearity, multi-dimensionality, mixed anatomic sampling, and possible autocorrelation of available immune data, it is challenging to identify the components of the innate and adaptive immune response that drive viral elimination. Novel mathematical models and analytical approaches are required to synthesize contemporaneously gathered cytokine, transcriptomic, flow cytometry, antibody response, and viral load data into a coherent story of viral control, and ultimately to discriminate drivers of mild versus severe infection.
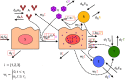
Methods: We investigated a dataset describing innate, SARS-CoV-2 specific T cell, and antibody responses in the lung during early and late stages of infection in immunologically naïve rhesus macaques. We used multi-model inference and ensemble modeling approaches from ecology and weather forecasting to compare and combine various competing models.
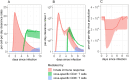
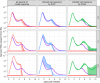
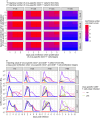
Results and discussion: Model outputs suggest that the innate immune response plays a crucial role in controlling early infection, while SARS-CoV-2 specific CD4+ T cells correspond to later viral elimination, and anti-spike IgG antibodies do not impact viral dynamics. Among the numerous genes potentially contributing to the innate response, we identified IFI27 as most closely linked to viral load decline. A 90% knockdown of the innate response from our validated model resulted in a ~10-fold increase in peak viral load during infection. Our approach provides a novel methodological framework for future analyses of similar complex, non-linear multi-component immunologic data sets.
Keywords: SARS-CoV-2; ensemble model; innate immunity; mathematical modeling; rhesus macaques; systems immunology; within-host infection dynamics.
Copyright © 2024 Byrne and Schiffer.
Conflict of interest statement
JS has done previous consultation for GSK and Pfizer. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets.Nat Microbiol. 2021 Jan;6(1):73-86. doi: 10.1038/s41564-020-00841-4. Epub 2020 Dec 18. Nat Microbiol. 2021. PMID: 33340034 Free PMC article.
-
Recovery from Acute SARS-CoV-2 Infection and Development of Anamnestic Immune Responses in T Cell-Depleted Rhesus Macaques.mBio. 2021 Aug 31;12(4):e0150321. doi: 10.1128/mBio.01503-21. Epub 2021 Jul 27. mBio. 2021. PMID: 34311582 Free PMC article.
-
Influence of Aerosol Delivered BCG Vaccination on Immunological and Disease Parameters Following SARS-CoV-2 Challenge in Rhesus Macaques.Front Immunol. 2022 Feb 9;12:801799. doi: 10.3389/fimmu.2021.801799. eCollection 2021. Front Immunol. 2022. PMID: 35222355 Free PMC article.
-
Immunological Aspects of SARS-CoV-2 Infection and the Putative Beneficial Role of Vitamin-D.Int J Mol Sci. 2021 May 16;22(10):5251. doi: 10.3390/ijms22105251. Int J Mol Sci. 2021. PMID: 34065735 Free PMC article. Review.
-
The immune response to SARS-CoV-2 and COVID-19 immunopathology - Current perspectives.Pulmonology. 2021 Sep-Oct;27(5):423-437. doi: 10.1016/j.pulmoe.2021.03.008. Epub 2021 Apr 9. Pulmonology. 2021. PMID: 33867315 Free PMC article. Review.
Cited by
-
A consensus mathematical model of vaccine-induced antibody dynamics for multiple vaccine platforms and pathogens.Front Immunol. 2025 Jun 26;16:1596518. doi: 10.3389/fimmu.2025.1596518. eCollection 2025. Front Immunol. 2025. PMID: 40642065 Free PMC article.
References
-
- Nelson CE, Namasivayam S, Foreman TW, Kauffman KD, Sakai S, Dorosky DE, et al. . NIAID/DIR Tuberculosis Imaging Program, Brooks K, Potter EL, et al. Mild SARS-CoV-2 infection in rhesus macaques is associated with viral control prior to antigen-specific T cell responses in tissues. Sci Immunol. (2022) 7:eabo0535. doi: 10.1126/sciimmunol.abo0535 - DOI - PMC - PubMed
-
- Chandrashekar A, Liu J, Yu J, McMahan K, Tostanoski LH, Jacob-Dolan C, et al. . Prior infection with SARS-CoV-2 WA1/2020 partially protects rhesus macaques against reinfection with B.1.1.7 and B.1.351 variants. Sci Transl Med. (2021) 13:eabj2641. doi: 10.1126/scitranslmed.abj2641 - DOI - PMC - PubMed
-
- Cohen KW, Linderman SL, Moodie Z, Czartoski J, Lai L, Mantus G, et al. . Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. (2021) 2:100354. doi: 10.1016/j.xcrm.2021.100354 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

