MICA and NKG2D gene polymorphisms influence graft survival, and response to therapy in kidney transplantation
- PMID: 39575256
- PMCID: PMC11578996
- DOI: 10.3389/fimmu.2024.1440887
MICA and NKG2D gene polymorphisms influence graft survival, and response to therapy in kidney transplantation
Abstract
Background: Antibody-mediated rejection is a significant cause of kidney transplant failure. Recent studies have shown that the MHC class I MICA gene influences the transplantation outcome. However, the role of the primary MICA receptor, NKG2D, has yet to be explored.
Aim: We aimed to investigate the correlation between recipient/donor MICA allele matching and NKG2D genotype with the risk of antibody-mediated rejection and their potential clinical effects and implications for organ maintenance therapy.
Methods: Of the 524 patients who underwent transplantation, 387 were eligible for the study. Complete MICA allele and two functional polymorphisms of NKG2D (rs1049174C>G and rs2255336G>A) were analyzed in 148 transplanted patients and 146 controls.
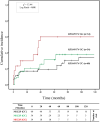
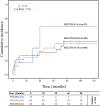
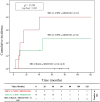
Results: Increased recipient/donor MICA allele mismatches correlate with an elevated risk of antibody-mediated rejection (X2 = 6.95; Log-rank=0.031). Notably, the rs1049174[GG] genotype contributes to a significantly increased risk of antibody-mediated rejection (X2 = 13.44; Log-rank=0.001 and X 2 = 0.34; Log-rank=0.84). The combined effect of two MICA allele mismatches and rs1049174[GG] genotype shows the highest risk (X2 = 23.21; Log-rank<0.001). Most importantly, patients with rs1049174[GG] and rs2255336[AA] genotypes may respond less to mTOR inhibitor immunosuppressive therapy than Calcineurin inhibitors (rs1049174[GG]; P=0.035; and rs2255336[AA]; P=0.002).
Conclusion: Recipient/donor MICA allele mismatches and specific NKG2D variants, as well as their combinations, influence kidney transplant outcomes, providing insights for personalized treatment and enhancing graft survival.
Keywords: DSA; MICA; NKG2D; antibody-mediated rejection; kidney transplant.
Copyright © 2024 Littera, Mocci, Argiolas, Littarru, Lai, Melis, Sanna, Mereu, Lorrai, Mascia, Angioi, Mascia, Matta, Lepori, Floris, Manieli, Bianco, Onnis, Rassu, Deidda, Carta, Giuressi, Perra, Chessa, Giglio and Pani.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, et al. . Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med. (1999) 341:1725–30. doi: 10.1056/NEJM199912023412303 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

