Identifying key underlying regulatory networks and predicting targets of orphan C/D box SNORD116 snoRNAs in Prader-Willi syndrome
- PMID: 39575480
- PMCID: PMC11662933
- DOI: 10.1093/nar/gkae1129
Identifying key underlying regulatory networks and predicting targets of orphan C/D box SNORD116 snoRNAs in Prader-Willi syndrome
Abstract
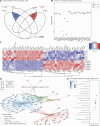
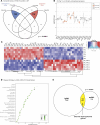
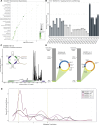
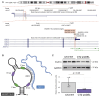
Prader-Willi syndrome (PWS) is a rare neurodevelopmental disorder characterized by neonatal hypotonia, followed by hyperphagia and obesity. Most PWS cases exhibit megabase-scale deletions of paternally imprinted 15q11-q13 locus. However, several PWS patients have been identified harboring much smaller deletions encompassing the SNORD116 gene cluster, suggesting these genes are direct drivers of PWS phenotypes. This cluster contains 30 copies of individual SNORD116 C/D box small nucleolar RNAs (snoRNAs). Many C/D box snoRNAs have been shown to guide chemical modifications of RNA molecules, often ribosomal RNA (rRNA). Conversely, SNORD116 snoRNAs show no significant complementarity to rRNA and their targets are unknown. Since many reported PWS cases lack their expression, it is crucial to identify the targets and functions of SNORD116. To address this we modeled PWS in two distinct human embryonic stem cell (hESC) lines with two different sized deletions, differentiated each into neurons, and compared differential gene expression. This analysis identified a novel set of 42 consistently dysregulated genes. These genes were significantly enriched for predicted SNORD116 targeting and we demonstrated impacts on FGF13 protein levels. Our results demonstrate the need for isogenic background comparisons and indicate a novel gene regulatory network controlled by SNORD116 is likely perturbed in PWS patients.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Update of
-
Identifying key underlying regulatory networks and predicting targets of orphan C/D box SNORD116 snoRNAs in Prader-Willi syndrome.bioRxiv [Preprint]. 2023 Oct 5:2023.10.03.560773. doi: 10.1101/2023.10.03.560773. bioRxiv. 2023. Update in: Nucleic Acids Res. 2024 Dec 11;52(22):13757-13774. doi: 10.1093/nar/gkae1129. PMID: 37873184 Free PMC article. Updated. Preprint.
References
-
- Prader A., Labhart A., Willi H.. Ein Syndrom von Adipositas, Kleinwuchs, Kryptorchismus und Oligophrenie nach myatonieartigem Zustand im Neugeborenenalter. Schweiz Med. Wochenschr. 1956; 86:1260–1261.
-
- Glenn C.C., Driscoll D.J., Yang T.P., Nicholls R.D.. Genomic imprinting: potential function and mechanisms revealed by the Prader-Willi and Angelman syndromes. Mol. Hum. Reprod. 1997; 3:321–332. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

