Shape transformations in peptide-DNA coacervates driven by enzyme-catalyzed deacetylation
- PMID: 39575590
- PMCID: PMC11582960
- DOI: 10.1039/d4sm01091d
Shape transformations in peptide-DNA coacervates driven by enzyme-catalyzed deacetylation
Abstract
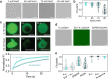
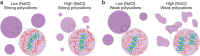
Biomolecular condensates formed by liquid-liquid phase separation (LLPS) are important organizers of biochemistry in living cells. Condensate formation can be dynamically regulated, for example, by protein binding or enzymatic processes. However, how enzymatic reactions can influence condensate shape and control shape transformations is less well understood. Here, we design a model condensate that can be formed by the enzymatic deacetylation of a small peptide by sirtuin-3 in the presence of DNA. Interestingly, upon nucleation condensates initially form gel-like aggregates that gradually transform into spherical droplets, displaying fusion and wetting. This process is governed by sirtuin-3 concentration, as more enzyme results in a faster aggregate-to-liquid transformation of the condensates. The counterintuitive transformation of gel-like to liquid-like condensates with increasing interaction strength between the peptide and DNA is recapitulated by forming condensates with different peptides and nucleic acids at increasing salt concentrations. Close to the critical point where coacervates dissolve, gel-like aggregates are formed with short double stranded DNA, but not with single stranded DNA or weakly binding peptides, even though the coacervate salt resistance is similar. At lower salt concentrations the interaction strength increases, and spherical, liquid-like condensates are formed. We attribute this behavior to bending of the DNA by oppositely charged peptides, which becomes stronger as the system moves further into the two-phase region. Overall, this work shows that enzymes can induce shape transformations of condensates and that condensate material properties do not necessarily reveal their stability.
Conflict of interest statement
There are no conflicts to declare.
Figures


References
-
- Yewdall N. A. André A. A. Lu T. Spruijt E. Curr. Opin. Colloid Interface Sci. 2021;52:101416. doi: 10.1016/j.cocis.2020.101416. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

