Laser dissection-assisted phloem transcriptomics highlights the metabolic and physiological changes accompanying clubroot disease progression in oilseed rape
- PMID: 39575835
- PMCID: PMC11703547
- DOI: 10.1111/tpj.17156
Laser dissection-assisted phloem transcriptomics highlights the metabolic and physiological changes accompanying clubroot disease progression in oilseed rape
Abstract
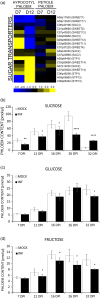
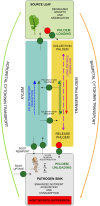
Plasmodiophora brassicae, a soil-borne biotroph, establishes galls as strong physiological sinks on Brassicaceae plants including Brassica napus and Arabidopsis thaliana. We compare transcriptional profiles of phloem dissected from leaf petioles and hypocotyls of healthy and infected B. napus plants. Our results highlight how pathogenesis accompanies phloem-mediated defence responses whilst exerting a strong influence on carbon-nitrogen (C-N) economy. We observe transcriptional changes indicating decreased aliphatic glucosinolate biosynthesis, fluctuating jasmonic acid responses, altered amino acid (AA) and nitrate transport, carbohydrate metabolism and modified cytokinin responses. Changes observed in phloem-dissected from upper versus lower plant organs point to phloem as a conduit in mediating C-N repartitioning, nutrition-related signalling and cytokinin dynamics over long distances during clubroot disease. To assess changes in physiology, we measured AAs, sugars and cytokinins, in phloem exudates from B. napus plants. Despite the decrease in most AA and sucrose levels, isopentyl-type cytokinins increased within infected plants. Furthermore, we employed Arabidopsis for visualising promoter activities of B. napus AA and N transporter orthologues and tested the impact of disrupted cytokinin transport during P. brassicae-induced gall formation using Atabcg14 mutants. Our physiological and microscopy studies show that the host developmental reaction to P. brassicae relies on cytokinin and is accompanied by intense nitrogen and carbon repartitioning. Overall, our work highlights the systemic aspects of host responses that should be taken into account when studying clubroot disease.
Keywords: Brassica napus; Plasmodiophora brassicae; clubroot; laser dissection transcriptomics; oilseed rape; phloem.
© 2024 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could appear to influence the work reported in this paper.
Figures





Similar articles
-
Comparative Transcriptome Analysis of Rutabaga (Brassica napus) Cultivars Indicates Activation of Salicylic Acid and Ethylene-Mediated Defenses in Response to Plasmodiophora brassicae.Int J Mol Sci. 2020 Nov 8;21(21):8381. doi: 10.3390/ijms21218381. Int J Mol Sci. 2020. PMID: 33171675 Free PMC article.
-
Hormonal Responses to Plasmodiophora brassicae Infection in Brassica napus Cultivars Differing in Their Pathogen Resistance.Int J Mol Sci. 2018 Dec 13;19(12):4024. doi: 10.3390/ijms19124024. Int J Mol Sci. 2018. PMID: 30551560 Free PMC article.
-
Transcriptome analysis of Brassica napus near-isogenic lines carrying clubroot resistance from turnip (Brassica rapa var. rapifera).Genome. 2025 Jan 1;68:1-17. doi: 10.1139/gen-2024-0182. Genome. 2025. PMID: 40479738
-
Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop.Mol Plant Pathol. 2012 Feb;13(2):105-13. doi: 10.1111/j.1364-3703.2011.00729.x. Epub 2011 Jun 1. Mol Plant Pathol. 2012. PMID: 21726396 Free PMC article. Review.
-
The clubroot pathogen Plasmodiophora brassicae: A profile update.Mol Plant Pathol. 2023 Feb;24(2):89-106. doi: 10.1111/mpp.13283. Epub 2022 Nov 29. Mol Plant Pathol. 2023. PMID: 36448235 Free PMC article. Review.
References
-
- Bieleski, R.L. (1964) The problem of halting enzyme action when extracting plant tissues. Analytical Biochemistry, 9, 431–442. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

