Cellulose degradation in Glenea cantor (Fabricius) (Coleoptera: Cerambycidae): functional characterization of GcEGaseZ7 and Cellulase reveals a novel enzymatic activity
- PMID: 39576188
- PMCID: PMC11583219
- DOI: 10.1093/jisesa/ieae101
Cellulose degradation in Glenea cantor (Fabricius) (Coleoptera: Cerambycidae): functional characterization of GcEGaseZ7 and Cellulase reveals a novel enzymatic activity
Erratum in
-
Correction to: Cellulose degradation in Glenea cantor (Fabricius) (Coleoptera: Cerambycidae): functional characterization of GcEGaseZ7 and Cellulase reveals a novel enzymatic activity.J Insect Sci. 2025 Mar 14;25(2):23. doi: 10.1093/jisesa/ieaf042. J Insect Sci. 2025. PMID: 40298837 Free PMC article. No abstract available.
Abstract
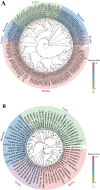
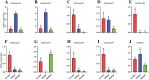
Glenea cantor (Fabricius) is an important forest pest that mainly attacks kapok trees, breaking down cellulose and lignin through 3 enzyme activities: endoglucanase, filter paper enzyme, and cellobiase. In this study, we unveiled the cloning and expression of 10 endoglucanase genes, GcEGase5A1, GcEGase5A2, GcEGaseZ2, GcEGaseZ3, GcEGaseZ4, GcEGaseZ5, GcEGaseZ7, GcEGaseZ8, GcEGaseZ9, and Cellulase, all of which exhibit enzymatic activities in G. cantor. These findings indicated that Cellulase shares sequence homology with beetle GHF45, whereas the other 9 endoglucanase genes are homologous to beetle GHF5. GcEGaseZ4 presented the highest expression in the foregut. In contrast, GcEGase5A2 and Cellulase presented peak expression in the midgut. Furthermore, GcEGaseZ7 was identified as the most highly expressed endoglucanase in the hindgut. Functional assays confirmed the ability of GcEGaseZ7 and Cellulase to degrade cellulose, and their cellulase activities were 75.57 ± 1.21 U/mg and 344.79 ± 6.91 U/mg, respectively. These results enhance our understanding of the complex cellulase system in insects and provide insights into the efficient digestion of cellulosic materials by wood-consuming insects. This research also has potential applications in bioenergy production and the development of biomaterials from lignocellulosic biomass.
Keywords: glycosyl hydrolase; insect enzymatic activity; longhorn beetles; prokaryotic expression.
© The Author(s) 2024. Published by Oxford University Press on behalf of Entomological Society of America.
Figures





References
-
- Amini E, Valls C, Yousefi H, et al. 2023. Ionic liquid/ZnO assisted preparation of high barrier cellulose nanocomposite films by in Situ Ringring-Opening polymerization of lactide monomers. J. Polym. Environ. 31(6):2576–2594. https://doi.org/10.1007/s10924-022-02740-7 - DOI
-
- Busch A, Kunert G, Heckel DG, et al. 2017. Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles. PLoS One 12(8):e0184305. https://doi.org/10.1371/journal.pone.0184305 - DOI - PMC - PubMed
-
- Busch A, Danchin EGJ, Pauchet Y. 2019. Functional diversification of horizontally acquired glycoside hydrolase family 45 (GH45) proteins in Phytophaga beetles. BMC Evol. Biol. 19(1):1–14. https://doi.org/10.1186/s12862-019-1429-9 - DOI - PMC - PubMed
-
- Busconi M, Berzolla A, Chiappini E. 2014. Preliminary data on cellulase encoding genes in the xylophagous beetle, Hylotrupes bajulus (Linnaeus). Int. Biodeterior. Biodegrad. 86(1):92–95. https://doi.org/10.1016/j.ibiod.2013.09.009 - DOI
-
- Chang QL, Sun JY, Xu YL, et al. 2006. Expression of a thermostable endoglucanase gene celD from Clostridium thermocellum in Escherichia coli and characterization of its recombinant enzyme. J. Agric. Biotechnol. 14(6):1000–1001. (In Chinese with English abstract).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

