AutoXAI4Omics: an automated explainable AI tool for omics and tabular data
- PMID: 39576223
- PMCID: PMC11583442
- DOI: 10.1093/bib/bbae593
AutoXAI4Omics: an automated explainable AI tool for omics and tabular data
Abstract
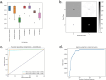
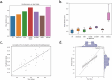
Machine learning (ML) methods offer opportunities for gaining insights into the intricate workings of complex biological systems, and their applications are increasingly prominent in the analysis of omics data to facilitate tasks, such as the identification of novel biomarkers and predictive modeling of phenotypes. For scientists and domain experts, leveraging user-friendly ML pipelines can be incredibly valuable, enabling them to run sophisticated, robust, and interpretable models without requiring in-depth expertise in coding or algorithmic optimization. By streamlining the process of model development and training, researchers can devote their time and energies to the critical tasks of biological interpretation and validation, thereby maximizing the scientific impact of ML-driven insights. Here, we present an entirely automated open-source explainable AI tool, AutoXAI4Omics, that performs classification and regression tasks from omics and tabular numerical data. AutoXAI4Omics accelerates scientific discovery by automating processes and decisions made by AI experts, e.g. selection of the best feature set, hyper-tuning of different ML algorithms and selection of the best ML model for a specific task and dataset. Prior to ML analysis AutoXAI4Omics incorporates feature filtering options that are tailored to specific omic data types. Moreover, the insights into the predictions that are provided by the tool through explainability analysis highlight associations between omic feature values and the targets under investigation, e.g. predicted phenotypes, facilitating the identification of novel actionable insights. AutoXAI4Omics is available at: https://github.com/IBM/AutoXAI4Omics.
Keywords: automated; explainable; machine learning; omics.
© The Author(s) 2024. Published by Oxford University Press.
Figures






References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

