Discovery of nostatin A, an azole-containing proteusin with prominent cytostatic and pro-apoptotic activity
- PMID: 39576263
- PMCID: PMC11583998
- DOI: 10.1039/d4ob01395f
Discovery of nostatin A, an azole-containing proteusin with prominent cytostatic and pro-apoptotic activity
Abstract
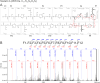
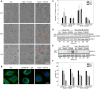
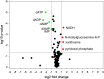
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are intriguing compounds with potential pharmacological applications. While many RiPPs are known as antimicrobial agents, a limited number of RiPPs with anti-proliferative effects in cancer cells are available. Here we report the discovery of nostatin A (NosA), a highly modified RiPP belonging among nitrile hydratase-like leader peptide RiPPs (proteusins), isolated from a terrestrial cyanobacterium Nostoc sp. Its structure was established based on the core peptide sequence encoded in the biosynthetic gene cluster recovered from the producing strain and subsequent detailed nuclear magnetic resonance and high-resolution mass spectrometry analyses. NosA, composed of a 30 amino-acid peptide core, features a unique combination of moieties previously not reported in RiPPs: the simultaneous presence of oxazole/thiazole heterocycles, dehydrobutyrine/dehydroalanine residues, and a sactionine bond. NosA includes an isobutyl-modified proline residue, highly unusual in natural products. NosA inhibits proliferation of multiple cancer cell lines at low nanomolar concentration while showing no hemolysis. It induces cell cycle arrest in S-phase followed by mitochondrial apoptosis employing a mechanism different from known tubulin binding and DNA damaging compounds. NosA also inhibits Staphylococcus strains while it exhibits no effect in other tested bacteria or yeasts. Due to its novel structure and selective bioactivity, NosA represents an excellent candidate for combinatorial chemistry approaches leading to development of novel NosA-based lead compounds.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
-
- Arnison P. G. Bibb M. J. Bierbaum G. Bowers A. A. Bugni T. S. Bulaj G. Camarero J. A. Campopiano D. J. Challis G. L. Clardy J. Cotter P. D. Craik D. J. Dawson M. Dittmann E. Donadio S. Dorrestein P. C. Entian K. D. Fischbach M. A. Garavelli J. S. Göransson U. Gruber C. W. Haft D. H. Hemscheidt T. K. Hertweck C. Hill C. Horswill A. R. Jaspars M. Kelly W. L. Klinman J. P. Kuipers O. P. Link A. J. Liu W. Marahiel M. A. Mitchell D. A. Moll G. N. Moore B. S. Müller R. Nair S. K. Nes I. F. Norris G. E. Olivera B. M. Onaka H. Patchett M. L. Piel J. Reaney M. J. T. Rebuffat S. Ross R. P. Sahl H. G. Schmidt E. W. Selsted M. E. Severinov K. Shen B. Sivonen K. Smith L. Stein T. Süssmuth R. D. Tagg J. R. Tang G. L. Truman A. W. Vederas J. C. Walsh C. T. Walton J. D. Wenzel S. C. Willey J. M. van der Donk W. A. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/C2NP20085F. - DOI - PMC - PubMed
-
- Cao L. Do T. Link A. J. Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs) J. Ind. Microbiol. Biotechnol. 2021;48 doi: 10.1093/jimb/kuab005. https://dx.doi.org/10.1093/jimb/kuab005 - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

