Utilizing a domain-specific large language model for LI-RADS v2018 categorization of free-text MRI reports: a feasibility study
- PMID: 39576290
- PMCID: PMC11584817
- DOI: 10.1186/s13244-024-01850-1
Utilizing a domain-specific large language model for LI-RADS v2018 categorization of free-text MRI reports: a feasibility study
Abstract
Objective: To develop a domain-specific large language model (LLM) for LI-RADS v2018 categorization of hepatic observations based on free-text descriptions extracted from MRI reports.
Material and methods: This retrospective study included 291 small liver observations, divided into training (n = 141), validation (n = 30), and test (n = 120) datasets. Of these, 120 were fictitious, and 171 were extracted from 175 MRI reports from a single institution. The algorithm's performance was compared to two independent radiologists and one hepatologist in a human replacement scenario, and considering two combined strategies (double reading with arbitration and triage). Agreement on LI-RADS category and dichotomic malignancy (LR-4, LR-5, and LR-M) were estimated using linear-weighted κ statistics and Cohen's κ, respectively. Sensitivity and specificity for LR-5 were calculated. The consensus agreement of three other radiologists served as the ground truth.
Results: The model showed moderate agreement against the ground truth for both LI-RADS categorization (κ = 0.54 [95% CI: 0.42-0.65]) and the dichotomized approach (κ = 0.58 [95% CI: 0.42-0.73]). Sensitivity and specificity for LR-5 were 0.76 (95% CI: 0.69-0.86) and 0.96 (95% CI: 0.91-1.00), respectively. When the chatbot was used as a triage tool, performance improved for LI-RADS categorization (κ = 0.86/0.87 for the two independent radiologists and κ = 0.76 for the hepatologist), dichotomized malignancy (κ = 0.94/0.91 and κ = 0.87) and LR-5 identification (1.00/0.98 and 0.85 sensitivity, 0.96/0.92 and 0.92 specificity), with no statistical significance compared to the human readers' individual performance. Through this strategy, the workload decreased by 45%.
Conclusion: LI-RADS v2018 categorization from unlabelled MRI reports is feasible using our LLM, and it enhances the efficiency of data curation.
Critical relevance statement: Our proof-of-concept study provides novel insights into the potential applications of LLMs, offering a real-world example of how these tools could be integrated into a local workflow to optimize data curation for research purposes.
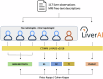
Key points: Automatic LI-RADS categorization from free-text reports would be beneficial to workflow and data mining. LiverAI, a GPT-4-based model, supported various strategies improving data curation efficiency by up to 60%. LLMs can integrate into workflows, significantly reducing radiologists' workload.
Keywords: Hepatocellular carcinoma; Natural language processing; Radiology; Report; Standardization.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study protocol was approved by the Clinical Research Ethics Committee from our institution (HCB/2023/0900). Informed consent was waived due to the retrospective character of data collection and the use of anonymized data. Consent for publication: Not applicable. Competing interests: The authors of this manuscript declare relationships with the following companies: J.R. has served as an advisor to Roche and has received lecture or consultancy fees from AstraZeneca and Roche. A.F. has received lecture fees from Gilead, Boston Scientific, Roche, AstraZeneca and MSD, also advisor fees from AstraZeneca, Roche, SIRTEX, Boston Scientific, AB Exact Science, Taiho and Guerbert. M.R. has served as an advisor and received lecture fees from AstraZeneca, Bayer, BMS, Eli Lilly, Geneos, Ipsen, Merck, Roche, Universal DX, and Engitix Therapeutics. Biotoscana Farma S.A. Travel support by: AstraZeneca, Roche, Bayer, BMS, Lilly. Ipsen Grant Research Support (to the institution): Bayer and Ipsen. Educational Support (to the institution): Bayer, AstraZeneca, BMS, Eisai- Merck MSD, Roche, Ipsen, Lilly, Terumo, Next, Boston Scientific, Ciscar Medical, and Eventy 03 LLC (Egypt). A.D.: has received speaker fees and travel grants from Bayer. The rest of the authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Figures




References
-
- RadReport Template Library (2020) Radiological Society of North America (2020). Available via https://radreport.org. Accessed 3 Jan 2023
-
- European Society of Radiology (2018) ESR paper on structured reporting in radiology. Insights Imaging. 10.1007/s13244-017-0588-8
-
- Chernyak V, Tang A, Do RK et al (2022) Liver imaging: it is time to adopt standardized terminology. Eur Radiol. 10.1007/s00330-022-08769-5 - PubMed
LinkOut - more resources
Full Text Sources

