Single-cell multi-omics sequencing uncovers region-specific plasticity of glioblastoma for complementary therapeutic targeting
- PMID: 39576855
- PMCID: PMC11584018
- DOI: 10.1126/sciadv.adn4306
Single-cell multi-omics sequencing uncovers region-specific plasticity of glioblastoma for complementary therapeutic targeting
Abstract
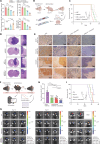
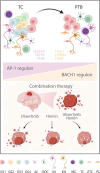
Glioblastoma (GBM) cells are highly heterogeneous and invasive, leading to treatment resistance and relapse. However, the molecular regulation in and distal to tumors remains elusive. Here, we collected paired tissues from the tumor core (TC) and peritumoral brain (PTB) for integrated snRNA-seq and snATAC-seq analyses. Tumor cells infiltrating PTB from TC behave more like oligodendrocyte progenitor cells than astrocytes at the transcriptome level. Dual-omics analyses further suggest that the distal regulatory regions in the tumor genome and specific transcription factors are potential determinants of regional heterogeneity. Notably, while activator protein 1 (AP-1) is active in all GBM states, its activity declines from TC to PTB, with another transcription factor, BACH1, showing the opposite trend. Combined inhibition of AP-1 and BACH1 more efficiently attenuates the tumor progression in mice and prolongs survival than either single-target treatment. Together, our work reveals marked molecular alterations of infiltrated GBM cells and a synergy of combination therapy targeting intratumor heterogeneity in and distal to GBM.
Figures





References
-
- Price M., Neff C., Nagarajan N., Kruchko C., Waite K. A., Cioffi G., Cordeiro B. B., Willmarth N., Penas-Prado M., Gilbert M. R., Armstrong T. S., Barnholtz-Sloan J. S., Ostrom Q. T., CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 26, iii1–iii53 (2024). - PMC - PubMed
-
- Stupp R., Mason W. P., Van Den Bent M. J., Weller M., Fisher B., Taphoorn M. J. B., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O.; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). - PubMed
-
- Milano M. T., Okunieff P., Donatello R. S., Mohile N. A., Sul J., Walter K. A., Korones D. N., Patterns and timing of recurrence after temozolomide-based chemoradiation for glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 78, 1147–1155 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

