Hemochromatosis neural archetype reveals iron disruption in motor circuits
- PMID: 39576859
- PMCID: PMC11584016
- DOI: 10.1126/sciadv.adp4431
Hemochromatosis neural archetype reveals iron disruption in motor circuits
Abstract
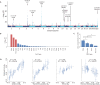
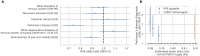
Our understanding of brain iron regulation and its disruption in disease is limited. Excess iron affects motor circuitry, contributing to Parkinson's disease (PD) risk. The molecular mechanisms regulating central iron levels, beyond a few well-known genes controlling peripheral iron, remain unclear. We generated scores based on the archetypal brain iron accumulation observed in magnetic resonance imaging scans of individuals with excessive dietary iron absorption and hemochromatosis risk. Genome-wide analysis revealed that this score is highly heritable, identifying loci associated with iron homeostasis, and driven by peripheral iron levels. Our score predicted gait abnormalities and showed a U-shaped relationship with PD risk, identifying individuals with threefold increased risk. These results establish a hormetic relationship between brain iron and PD risk, where central iron levels are strongly determined by genetics via peripheral iron. This framework combining forward and reverse genetics is a powerful study design to understand genomic drivers underlying high dimensional phenotypes.
Figures

References
-
- Rouault T. A., Iron metabolism in the CNS: Implications for neurodegenerative diseases. Nat. Rev. Neurosci. 14, 551–564 (2013). - PubMed
-
- Pasricha S.-R., Tye-Din J., Muckenthaler M. U., Swinkels D. W., Iron deficiency. Lancet 397, 233–248 (2021). - PubMed
-
- Do Van B., Gouel F., Jonneaux A., Timmerman K., Gelé P., Pétrault M., Bastide M., Laloux C., Moreau C., Bordet R., Devos D., Devedjian J.-C., Ferroptosis, a newly characterized form of cell death in Parkinson’s disease that is regulated by PKC. Neurobiol. Dis. 94, 169–178 (2016). - PubMed

