Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens
- PMID: 39576860
- PMCID: PMC11584006
- DOI: 10.1126/sciadv.adq6150
Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens
Abstract
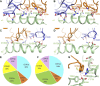
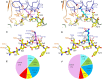
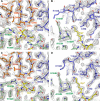
T cell receptors (TCRs) that recognize cancer neoantigens are important for anticancer immune responses and immunotherapy. Understanding the structural basis of TCR recognition of neoantigens provides insights into their exquisite specificity and can enable design of optimized TCRs. We determined crystal structures of a human TCR in complex with NRAS Q61K and Q61R neoantigen peptides and HLA-A1 major histocompatibility complex (MHC), revealing the molecular underpinnings for dual recognition and specificity versus wild-type NRAS peptide. We then used multiple versions of AlphaFold to model the corresponding complex structures, given the challenge of immune recognition for such methods. One implementation of AlphaFold2 (TCRmodel2) with additional sampling was able to generate accurate models of the complexes, while AlphaFold3 also showed strong performance, although success was lower for other complexes. This study provides insights into TCR recognition of a shared cancer neoantigen as well as the utility and practical considerations for using AlphaFold to model TCR-peptide-MHC complexes.
Figures


Update of
-
Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.bioRxiv [Preprint]. 2024 May 23:2024.05.21.595215. doi: 10.1101/2024.05.21.595215. bioRxiv. 2024. Update in: Sci Adv. 2024 Nov 22;10(47):eadq6150. doi: 10.1126/sciadv.adq6150. PMID: 38826362 Free PMC article. Updated. Preprint.
References
-
- Zacharakis N., Chinnasamy H., Black M., Xu H., Lu Y. C., Zheng Z., Pasetto A., Langhan M., Shelton T., Prickett T., Gartner J., Jia L., Trebska-McGowan K., Somerville R. P., Robbins P. F., Rosenberg S. A., Goff S. L., Feldman S. A., Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24, 724–730 (2018). - PMC - PubMed
-
- Stevanovic S., Pasetto A., Helman S. R., Gartner J. J., Prickett T. D., Howie B., Robins H. S., Robbins P. F., Klebanoff C. A., Rosenberg S. A., Hinrichs C. S., Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 356, 200–205 (2017). - PMC - PubMed
-
- Tran E., Turcotte S., Gros A., Robbins P. F., Lu Y. C., Dudley M. E., Wunderlich J. R., Somerville R. P., Hogan K., Hinrichs C. S., Parkhurst M. R., Yang J. C., Rosenberg S. A., Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

