LTβR deficiency causes lymph node aplasia and impaired B cell differentiation
- PMID: 39576873
- PMCID: PMC7618087
- DOI: 10.1126/sciimmunol.adq8796
LTβR deficiency causes lymph node aplasia and impaired B cell differentiation
Abstract
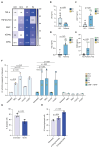
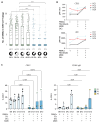
Secondary lymphoid organs (SLOs) provide the confined microenvironment required for stromal cells to interact with immune cells to initiate adaptive immune responses resulting in B cell differentiation. Here, we studied three patients from two families with functional hyposplenism, absence of tonsils, and complete lymph node aplasia, leading to recurrent bacterial and viral infections. We identified biallelic loss-of-function mutations in LTBR, encoding the lymphotoxin beta receptor (LTβR), primarily expressed on stromal cells. Patients with LTβR deficiency had hypogammaglobulinemia, diminished memory B cells, regulatory and follicular T helper cells, and dysregulated expression of several tumor necrosis factor family members. B cell differentiation in an ex vivo coculture system was intact, implying that the observed B cell defects were not intrinsic in nature and instead resulted from LTβR-dependent stromal cell interaction signaling critical for SLO formation. Collectively, we define a human inborn error of immunity caused primarily by a stromal defect affecting the development and function of SLOs.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures



References
-
- Horsnell HL, Tetley RJ, De Belly H, Makris S, Millward LJ, Benjamin AC, Heeringa L, de Winde CM, Paluch EK, Mao Y, Acton SE. Lymph node homeostasis and adaptation to immune challenge resolved by fibroblast network mechanics. Nat Immunol. 2022;23:1169–1182. doi: 10.1038/s41590-022-01272-5. - DOI - PMC - PubMed
-
- Cruz de Casas P, Knopper K, Dey Sarkar R, Kastenmuller W. Same yet different - how lymph node heterogeneity affects immune responses. Nat Rev Immunol. 2023 - PubMed
-
- Victora GD, Nussenzweig MC. Germinal Centers. Annu Rev Immunol. 2022;40:413–442. - PubMed
-
- Vetrie D, Vorechovsky I, Sideras P, Holland J, Davies A, Flinter F, Hammarstrom L, Kinnon C, Levinsky R, Bobrow M, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361:226–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

