The 1000 Chinese Indigenous Pig Genomes Project provides insights into the genomic architecture of pigs
- PMID: 39578420
- PMCID: PMC11584710
- DOI: 10.1038/s41467-024-54471-z
The 1000 Chinese Indigenous Pig Genomes Project provides insights into the genomic architecture of pigs
Abstract
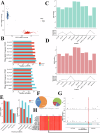
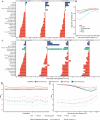
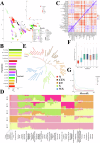
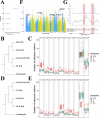
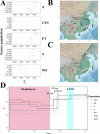
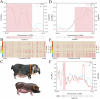
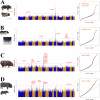
Pigs play a central role in human livelihoods in China, but a lack of systematic large-scale whole-genome sequencing of Chinese domestic pigs has hindered genetic studies. Here, we present the 1000 Chinese Indigenous Pig Genomes Project sequencing dataset, comprising 1011 indigenous individuals from 50 pig populations covering approximately two-thirds of China's administrative divisions. Based on the deep sequencing (~25.95×) of these pigs, we identify 63.62 million genomic variants, and provide a population-specific reference panel to improve the imputation performance of Chinese domestic pig populations. Using a combination of methods, we detect an ancient admixture event related to a human immigration climax in the 13th century, which may have contributed to the formation of southeast-central Chinese pig populations. Analyzing the haplotypes of the Y chromosome shows that the indigenous populations residing around the Taihu Lake Basin exhibit a unique haplotype. Furthermore, we find a 13 kb region in the THSD7A gene that may relate to high-altitude adaptation, and a 0.47 Mb region on chromosome 7 that is significantly associated with body size traits. These results highlight the value of our genomic resource in facilitating genomic architecture and complex traits studies in pigs.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
-
- Frantz, L. A. F., Madsen, O., Megens, H. J., Groenen, M. A. M. & Lohse, K. Testing models of speciation from genome sequences: divergence and asymmetric admixture in Island South-East Asian Sus species during the Plio-Pleistocene climatic fluctuations. Mol. Ecol.23, 5566–5574 (2014). - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

