Viral sequence determines HLA-E-restricted T cell recognition of hepatitis B surface antigen
- PMID: 39578466
- PMCID: PMC11584656
- DOI: 10.1038/s41467-024-54378-9
Viral sequence determines HLA-E-restricted T cell recognition of hepatitis B surface antigen
Abstract
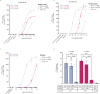
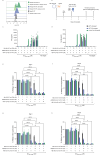
The non-polymorphic HLA-E molecule offers opportunities for new universal immunotherapeutic approaches to chronic infectious diseases. Chronic Hepatitis B virus (HBV) infection is driven in part by T cell dysfunction due to elevated levels of the HBV envelope (Env) protein hepatitis B surface antigen (HBsAg). Here we report the characterization of three genotypic variants of an HLA-E-binding HBsAg peptide, Env371-379, identified through bioinformatic predictions and verified by biochemical and cellular assays. Using a soluble affinity-enhanced T cell receptor (TCR) (a09b08)-anti-CD3 bispecific molecule to probe HLA-E presentation of the Env371-379 peptides, we demonstrate that only the most stable Env371-379 variant, L6I, elicits functional responses to a09b08-anti-CD3-redirected polyclonal T cells co-cultured with targets expressing endogenous HBsAg. Furthermore, HLA-E-Env371-379 L6I-specific CD8+ T cells are detectable in HBV-naïve donors and people with chronic HBV after in vitro priming. In conclusion, we provide evidence for HLA-E-mediated HBV Env peptide presentation, and highlight the effect of viral mutations on the stability and targetability of pHLA-E molecules.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: G.M., R.L.P., R.K., V.I., R.J.S., M.M.C., V.K., R.P., A.J., J.D., T.H., W.B., G.P., K.O., D.K., A.S., C.B., R.R., C.P., T.G., A.T., A.W., M.D., D.G., M.H., R.J.C., K.E.A., L.D., A.K., S.L., M.S., and L.F.G. are or were employees of Immunocore Ltd.
Figures





References
-
- WHO. Hepatitis B. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (2024).
-
- Jeng, W.-J., Papatheodoridis, G. V. & Lok, A. S. F. Hepatitis B. Lancet401, 1039–1052 (2023). - PubMed
-
- Xie, Y. Infectious agents associated cancers: epidemiology and molecular biology. Adv. Exp. Med. Biol.1018, 11–21 (2017). - PubMed
-
- Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.68, 394–424 (2018). - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

