Mitochondrial heteroplasmy improves risk prediction for myeloid neoplasms
- PMID: 39578475
- PMCID: PMC11584845
- DOI: 10.1038/s41467-024-54443-3
Mitochondrial heteroplasmy improves risk prediction for myeloid neoplasms
Abstract
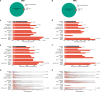
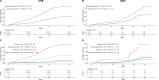
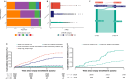
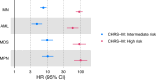
Clonal hematopoiesis of indeterminate potential is the primary pathogenic risk factor for myeloid neoplasms, while heteroplasmy (mutations in a subset of cellular mitochondrial DNA) is another marker of clonal expansion associated with hematological malignancies. We explore how these two markers relate and influence myeloid neoplasms incidence, and their role in risk stratification. We find that heteroplasmy is more common in individuals with clonal hematopoiesis of indeterminate potential, particularly those with higher variant allele fractions, multiple mutations, or spliceosome machinery mutations. Individuals with both markers have a higher risk of myeloid neoplasms than those with either alone. Furthermore, heteroplasmic variants with higher predicted deleteriousness increase the risk of myeloid neoplasms. Incorporating heteroplasmy in an existing risk score model for individuals with clonal hematopoiesis of indeterminate potential significantly improves sensitivity and better identifies high-risk groups. This suggests heteroplasmy as a clonal expansion marker and potentially as a biomarker for myeloid neoplasms development.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Lin, J. S., Perdue, L. A., Henrikson, N. B., Bean, S. I. & Blasi, P. R. Screening for Colorectal Cancer: An Evidence Update for the U.S. Preventive Services Task Force. (Agency for Healthcare Research and Quality (US), Rockville (MD), 2021). - PubMed
-
- Jonas, D. E. et al. Screening for Lung Cancer With Low-Dose Computed Tomography: An Evidence Review for the U.S. Preventive Services Task Force. (Agency for Healthcare Research and Quality (US), Rockville (MD), 2021). - PubMed
-
- Melnikow, J., Fenton, J. J., Miglioretti, D., Whitlock, E. P. & Weyrich, M. S. Screening for Breast Cancer With Digital Breast Tomosynthesis. (Agency for Healthcare Research and Quality (US), Rockville (MD), 2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL156144/HL/NHLBI NIH HHS/United States
- R01HL156144/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL144569/HL/NHLBI NIH HHS/United States
- 75N92022D00001/HL/NHLBI NIH HHS/United States
- 75N92022D00002/HL/NHLBI NIH HHS/United States
- 75N92022D00003/HL/NHLBI NIH HHS/United States
- 75N92022D00004/HL/NHLBI NIH HHS/United States
- 75N92022D00005/HL/NHLBI NIH HHS/United States
- U01 CA164975/CA/NCI NIH HHS/United States
- P01 CA265748/CA/NCI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- HHSN268201500015C/HL/NHLBI NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- HHSN268201000021C/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- HHSN268201000001I/HL/NHLBI NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- UM1 HG008898/HG/NHGRI NIH HHS/United States
- R01 HL131136/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

