Targeting heterochromatin eliminates chronic myelomonocytic leukemia malignant stem cells through reactivation of retroelements and immune pathways
- PMID: 39578583
- PMCID: PMC11584673
- DOI: 10.1038/s42003-024-07214-1
Targeting heterochromatin eliminates chronic myelomonocytic leukemia malignant stem cells through reactivation of retroelements and immune pathways
Abstract
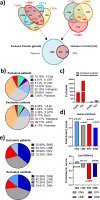
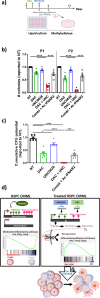
Chronic myelomonocytic leukemia (CMML) is a severe myeloid malignancy affecting the elderly, for which therapeutic options are limited. DNA hypomethylating agents (HMAs) provide transient responses, failing to eradicate the malignant clone. Hematopoietic stem cell (HSC) aging involves heterochromatin reorganization, evidenced by alterations in histone marks H3K9me2 and H3K9me3. These repressive marks together with DNA methylation are essential for suppressing transposable elements (TEs). In solid cancers, the antitumor efficacy of HMAs involves the derepression of TEs, mimicking a state of viral infection. In this study, we demonstrate a significant disorganization of heterochromatin in CMML HSCs and progenitors (HSPCs) characterized by an increase in the repressive mark H3K9me2, mainly at the level of TEs, and a repression of immune and age-associated transcripts. Combining HMAs with G9A/GLP H3K9me2 methyltransferase inhibitors reactivates these pathways, selectively targeting mutated cells while preserving wild-type HSCs, thus offering new therapeutic avenues for this severe myeloid malignancy.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: E.T. is an Editorial Board Member for Communications Biology, but was not involved in the editorial review of, nor the decision to publish this article. The other authors declare no competing interests.
Figures








References
-
- Itzykson, R. et al. Clonal architecture of chronic myelomonocytic leukemias. Blood121, 2186–2198 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

