CelloType: a unified model for segmentation and classification of tissue images
- PMID: 39578628
- PMCID: PMC11810770
- DOI: 10.1038/s41592-024-02513-1
CelloType: a unified model for segmentation and classification of tissue images
Abstract
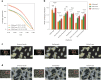
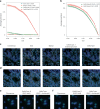
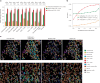
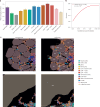
Cell segmentation and classification are critical tasks in spatial omics data analysis. Here we introduce CelloType, an end-to-end model designed for cell segmentation and classification for image-based spatial omics data. Unlike the traditional two-stage approach of segmentation followed by classification, CelloType adopts a multitask learning strategy that integrates these tasks, simultaneously enhancing the performance of both. CelloType leverages transformer-based deep learning techniques for improved accuracy in object detection, segmentation and classification. It outperforms existing segmentation methods on a variety of multiplexed fluorescence and spatial transcriptomic images. In terms of cell type classification, CelloType surpasses a model composed of state-of-the-art methods for individual tasks and a high-performance instance segmentation model. Using multiplexed tissue images, we further demonstrate the utility of CelloType for multiscale segmentation and classification of both cellular and noncellular elements in a tissue. The enhanced accuracy and multitask learning ability of CelloType facilitate automated annotation of rapidly growing spatial omics data.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
CelloType: A Unified Model for Segmentation and Classification of Tissue Images.bioRxiv [Preprint]. 2024 Sep 19:2024.09.15.613139. doi: 10.1101/2024.09.15.613139. bioRxiv. 2024. Update in: Nat Methods. 2025 Feb;22(2):348-357. doi: 10.1038/s41592-024-02513-1. PMID: 39345491 Free PMC article. Updated. Preprint.
References
-
- He, K., Zhang, X., Ren, S. & Sun, J. I. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

