A dual typing system establishment and global diversity analysis for sapoviruses
- PMID: 39578768
- PMCID: PMC11583745
- DOI: 10.1186/s12864-024-11048-w
A dual typing system establishment and global diversity analysis for sapoviruses
Abstract
Background: The genus Sapovirus in the family Caliciviridae comprises of a genetically diverse group of viruses that are responsible for causing acute gastroenteritis in both human and animals globally. As the number of sequences continues to grow and more recombinant sequences are identified, the classification criteria of genogroups and genotypes of sapovirus need to be further refined. In this study, we aimed to optimize the classification of sapoviruses.
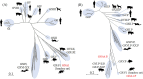
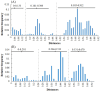
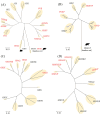
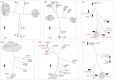
Results: Through evolutionary clustering and genetic distance analysis, we have updated the classification criteria for VP1 genogroup and genotypes. We adjusted the original mean values ± 3 standard deviations (SD) of genetic distances to mean values ± 2.5SD, resulting the corresponding cutoff values for the same genotype and genogroup set at <0.161 and <0.503, respectively. Additionally, we established classification criteria for RdRp types and groups, referred to as P-types and P-groups,, with mean values ± 2SD and cutoff values of <0.266 and <0.531 for the same type and group, respectively. This refinement has expanded the VP1 genogroups to thirty-four and identified twenty-four P-groups. For human sapoviruses, the new criteria have resulted in the addition of one genotype, GV.PNA1. Moreover, the new criteria defined three P-groups and 21 P-types for human sapoviruses. Spatial-temporal analysis revealed no specific distribution pattern for human sapoviruses.
Conclusions: We established a dual typing system on classification based on VP1 and RdRp nucleotide sequences for sapoviruses.
Keywords: Classification; Dual-typing; Genetic distance; Genogroup; Genotype; Sapoviruses.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

