Primary cartilage transcriptional signatures reflect cell-type-specific molecular pathways underpinning osteoarthritis
- PMID: 39579762
- PMCID: PMC11639091
- DOI: 10.1016/j.ajhg.2024.10.019
Primary cartilage transcriptional signatures reflect cell-type-specific molecular pathways underpinning osteoarthritis
Abstract
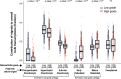
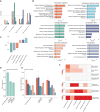
Translational efforts in osteoarthritis are hampered by a gap in our understanding of disease processes at the molecular level. Here, we present evidence of pronounced transcriptional changes in high- and low-disease-grade cartilage tissue, pointing to embryonic processes involved in disease progression. We identify shared transcriptional programs between osteoarthritis cartilage and cell populations in the human embryonic and fetal limb, pointing to increases in pre-hypertrophic chondrocytes' transcriptional programs in low-grade cartilage and increases in osteoblastic signatures in high-grade disease tissue. We find that osteoarthritis genetic risk signals are enriched in six gene co-expression modules and show that these transcriptional signatures reflect cell-type-specific expression along the endochondral ossification developmental trajectory. Using this network approach in combination with causal inference analysis, we present evidence of a causal effect on osteoarthritis risk for variants associated with the expression of ten genes that have not been previously reported as effector genes in genome-wide association studies in osteoarthritis. Our findings point to key molecular pathways as drivers of cartilage degeneration and identify high-value drug targets and repurposing opportunities.
Keywords: RNA sequencing; cell-type-specific expression; chondrocyte populations; disease pathways; endochondral ossification; gene co-expression networks; gene set analysis; osteoarthritis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests In the last three years, S.A.T. has been a remunerated consultant for Sanofi, Foresite Labs. and Qiagen and is a consultant and equity holder of TransitionBio and EnsoCell Therapeutics.
Figures





References
-
- Vos T., Lim S.S., Abbafati C., Abbas K.M., Abbasi M., Abbasifard M., Abbasi-Kangevari M., Abbastabar H., Abd-Allah F., Abdelalim A., et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–1222. doi: 10.1016/S0140-6736(20)30925-9. - DOI - PMC - PubMed
-
- Arden N.K., Perry T.A., Bannuru R.R., Bruyère O., Cooper C., Haugen I.K., Hochberg M.C., McAlindon T.E., Mobasheri A., Reginster J.-Y. Non-surgical management of knee osteoarthritis: comparison of ESCEO and OARSI 2019 guidelines. Nat. Rev. Rheumatol. 2021;17:59–66. doi: 10.1038/s41584-020-00523-9. - DOI - PubMed
-
- Boer C.G., Hatzikotoulas K., Southam L., Stefánsdóttir L., Zhang Y., Coutinho de Almeida R., Wu T.T., Zheng J., Hartley A., Teder-Laving M., et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell. 2021;184:4784–4818.e17. doi: 10.1016/j.cell.2021.07.038. - DOI - PMC - PubMed
-
- Rice S.J., Tselepi M., Sorial A.K., Aubourg G., Shepherd C., Almarza D., Skelton A.J., Pangou I., Deehan D., Reynard L.N., Loughlin J. Prioritization of PLEC and GRINA as Osteoarthritis Risk Genes Through the Identification and Characterization of Novel Methylation Quantitative Trait Loci. Arthritis Rheumatol. 2019;71:1285–1296. doi: 10.1002/art.40849. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

