Spatial dissection of tumour microenvironments in gastric cancers reveals the immunosuppressive crosstalk between CCL2+ fibroblasts and STAT3-activated macrophages
- PMID: 39580151
- PMCID: PMC12013559
- DOI: 10.1136/gutjnl-2024-332901
Spatial dissection of tumour microenvironments in gastric cancers reveals the immunosuppressive crosstalk between CCL2+ fibroblasts and STAT3-activated macrophages
Abstract
Background: A spatially resolved, niche-level analysis of tumour microenvironments (TME) can provide insights into cellular interactions and their functional impacts in gastric cancers (GC).
Objective: Our goal was to translate the spatial organisation of GC ecosystems into a functional landscape of cellular interactions involving malignant, stromal and immune cells.
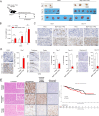
Design: We performed spatial transcriptomics on nine primary GC samples using the Visium platform to delineate the transcriptional landscape and dynamics of malignant, stromal and immune cells within the GC tissue architecture, highlighting cellular crosstalks and their functional consequences in the TME.
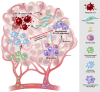
Results: GC spatial transcriptomes with substantial cellular heterogeneity were delineated into six regional compartments. Specifically, the fibroblast-enriched TME upregulates epithelial-to-mesenchymal transformation and immunosuppressive response in malignant and TME cells, respectively. Cell type-specific transcriptional dynamics revealed that malignant and endothelial cells promote the cellular proliferations of TME cells, whereas the fibroblasts and immune cells are associated with procancer and anticancer immunity, respectively. Ligand-receptor analysis revealed that CCL2-expressing fibroblasts promote the tumour progression via JAK-STAT3 signalling and inflammatory response in tumour-infiltrated macrophages. CCL2+ fibroblasts and STAT3-activated macrophages are co-localised and their co-abundance was associated with unfavourable prognosis. We experimentally validated that CCL2+ fibroblasts recruit myeloid cells and stimulate STAT3 activation in recruited macrophages. The development of immunosuppressive TME by CCL2+ fibroblasts were also validated in syngeneic mouse models.
Conclusion: GC spatial transcriptomes revealed functional cellular crosstalk involving multiple cell types among which the interaction between CCL2+ fibroblasts and STAT3-activated macrophages plays roles in establishing immune-suppressive GC TME with potential clinical relevance.
Keywords: gastric cancer.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures





References
-
- Bang Y-J, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
