Molecular insights into AGS3's role in spindle orientation: a biochemical perspective
- PMID: 39580365
- PMCID: PMC12151148
- DOI: 10.1093/jmcb/mjae049
Molecular insights into AGS3's role in spindle orientation: a biochemical perspective
Abstract
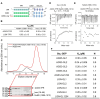
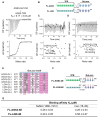
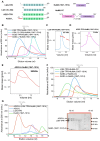
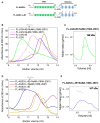
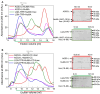
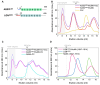
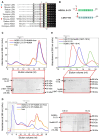
The intrinsic regulation of spindle orientation during asymmetric cell division depends on the evolutionarily conserved protein complex LGN (Pins)/NuMA (Mud)/Gα⋅GDP. While the role of LGN and its Drosophila orthologue Pins is well-established, the function of AGS3, the paralogue of LGN, in spindle orientation during cell division remains controversial. This study substantiates the contentious nature of AGS3's function through systematic biochemical characterizations. The results confirm the high conservation of AGS3 in its functional structural domains, similar to LGN, and its comparable ability to bind to partners including NuMA, Insc, and Gαi3⋅GDP. However, in contrast to LGN, AGS3 and the microtubule-binding protein NuMA are unable to form stable hetero-hexamers or higher-order oligomeric complexes that are pivotal for effective regulation of spindle orientation. It was found that this notable difference between AGS3 and LGN stems from the N-terminal sequence preceding the conserved TPR motifs, which spans ∼20 residues. Furthermore, our findings substantiate the disruptive effect of Insc on the oligomeric AGS3/NuMA complex, while showing no impact on the oligomeric LGN/NuMA complex. Consequently, Insc emerges as an additional regulatory factor that distinguishes the functional roles of AGS3 and LGN, leading to the impairment of AGS3's ability to actively reorient the mitotic spindle. These results elucidate the molecular basis underlying the observed functional disparity in spindle orientation between LGN and AGS3, providing valuable insights into the regulation of cell division at the molecular level.
Keywords: AGS3; LGN; NuMA; asymmetric cell division; spindle orientation.
© The Author(s) (2024). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, CEMCS, CAS.
Figures

References
-
- Adhikari A., Sprang S.R. (2003). Thermodynamic characterization of the binding of activator of G protein signaling 3 (AGS3) and peptides derived from AGS3 with Gα. J. Biol. Chem. 278, 51825–51832. - PubMed
-
- Bernard M.L., Peterson Y.K., Chung P. et al. (2001). Selective interaction of AGS3 with G-proteins and the influence of AGS3 on the activation state of G-proteins. J. Biol. Chem. 276, 1585–1593. - PubMed
-
- Blumer J.B., Oner S.S., Lanier S.M. (2012). Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol. 204, 202–218. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

