Genome-wide methylation profiling differentiates benign from aggressive and metastatic pituitary neuroendocrine tumors
- PMID: 39580368
- PMCID: PMC11585505
- DOI: 10.1007/s00401-024-02836-5
Genome-wide methylation profiling differentiates benign from aggressive and metastatic pituitary neuroendocrine tumors
Erratum in
-
Correction: Genome‑wide methylation profiling differentiates benign from aggressive and metastatic pituitary neuroendocrine tumors.Acta Neuropathol. 2025 Jan 2;149(1):4. doi: 10.1007/s00401-024-02838-3. Acta Neuropathol. 2025. PMID: 39747763 Free PMC article. No abstract available.
Abstract
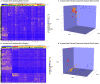
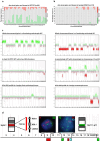
Aggressive pituitary neuroendocrine tumors (PitNETs)/adenomas are characterized by progressive growth despite surgery and all standard medical therapies and radiotherapy. A subset will metastasize to the brain and/or distant locations and are termed metastatic PitNETs (pituitary carcinomas). Studies of potential prognostic markers have been limited due to the rarity of these tumors. A few recurrent somatic mutations have been identified, and epigenetic alterations and chromosomal rearrangements have not been explored in larger cohorts of aggressive and metastatic PitNETs. In this study, we performed genome-wide methylation analysis, including copy-number variation (CNV) calculations, on tumor tissue specimens from a large international cohort of 64 patients with aggressive (48) and metastatic (16) pituitary tumors. Twelve patients with non-invasive pituitary tumors (Knosp 0-2) exhibiting an indolent course over a 5 year follow-up served as controls. In an unsupervised hierarchical cluster analysis, aggressive/metastatic PitNETs clustered separately from benign pituitary tumors, and, when only specimens from the first surgery were analyzed, three separate clusters were identified: aggressive, metastatic, and benign PitNETs. Numerous CNV events affecting chromosomal arms and whole chromosomes were frequent in aggressive and metastatic, whereas benign tumors had normal chromosomal copy numbers with only few alterations. Genome-wide methylation analysis revealed different CNV profiles and a clear separation between aggressive/metastatic and benign pituitary tumors, potentially providing biomarkers for identification of these tumors with a worse prognosis at the time of first surgery. The data may refine follow-up routines and contribute to the timely introduction of adjuvant therapy in patients harboring, or at risk of developing, aggressive or metastatic pituitary tumors.
Keywords: Aggressive pituitary tumor; Methylation analysis; Pituitary adenoma; Pituitary carcinoma; Pituitary neuroendocrine tumor.
© 2024. The Author(s).
Figures



References
-
- Caimari F, Korbonits M (2016) Novel genetic causes of pituitary adenomas. Clin Cancer Res 22:5030–5042. 10.1158/1078-0432.CCR-16-0452 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

