Single-cell RNA sequencing reveals immune microenvironment niche transitions during the invasive and metastatic processes of ground-glass nodules and part-solid nodules in lung adenocarcinoma
- PMID: 39580469
- PMCID: PMC11585206
- DOI: 10.1186/s12943-024-02177-7
Single-cell RNA sequencing reveals immune microenvironment niche transitions during the invasive and metastatic processes of ground-glass nodules and part-solid nodules in lung adenocarcinoma
Abstract
Background: Radiographically, ground-glass nodules (GGN) and part-solid nodules (PSN) in lung adenocarcinoma (LUAD) have significant heterogeneity in their clinical manifestations, biological characteristics, and prognosis. This study aimed to explore the heterogeneity of LUAD in different radiological phenotypes and associated factors influencing tumor evolution.
Methods: We performed single-cell RNA sequencing (scRNA-seq) on tumor tissues from eight and seven cases of GGN- and PSN-LUAD, respectively, at different disease stages, including minimally invasive adenocarcinoma (MIA), invasive adenocarcinoma (IAC), and metastatic lung cancer (MLC). Additionally, we analyzed adjacent normal tissues from four cases. Immunohistochemistry, multiplex immunofluorescence, and external scRNA-seq data were employed to confirm the expression of signature genes as well as the distribution patterns of CXCL9 + TAMs and TREM2 + TAMs. A LUAD mouse model was generated using gene editing, organoid culture, and orthotopic transplantation techniques, and comprehensive analyses such as histopathology, RNA sequencing, and Western blotting were performed to validate key pathways.
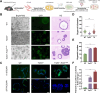
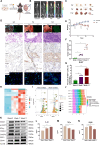
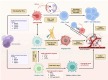
Results: Diverse cellular compositions were observed in the tumor microenvironment (TME) during GGN- and PSN-LUAD invasion and metastasis. Notably, CXCL9 + and TREM2 + tumor-associated macrophages (TAMs) exhibited the most significant enrichment changes. It was found that GGN-LUAD exhibited a stronger immune response than PSN-LUAD, with increased interaction between CXCL9 + TAMs and CD8 + tissue-resident memory T cells during invasion stage (MIA-IAC). Conversely, greater interactions between TREM2 + TAMs and tumor cells were observed in PSN-LUAD during the MLC stage. Additionally, TREM2 + TAMs were found to differentiate into TREM2 + /SPP1 + and TREM2 + /SPP1- TAMs at different stages, which promotes tumor progression. This study also emphasizes that during the transdifferentiation process of GGN- and PSN-LUAD, IFN-γ activates the STAT1 signaling pathway to regulate the activation of CXCL9 + TAMs, and further recruiting CD8 + Trm cells and activating T cells through MHC class I antigen presentation. The role of the IFN-γ/STAT1 pathway in the occurrence and development of LUAD was further validated by animal experiments.
Conclusions: Our findings offer a potential therapeutic strategy to maintain a dynamic balance within the TME and improve the immunotherapy efficacy by modulating the relative proportions and functional states of CXCL9 + TAMs and TREM2 + TAMs.
Keywords: CXCL9; Lung adenocarcinoma; Pulmonary nodule; SPP1; Single-cell RNA sequencing; TREM2; Tumor microenvironment; Tumor-associated macrophage.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study adhered to the principles of the Declaration of Helsinki and was approved by the Institutional Review Boards of the Chengdu University of Traditional Chinese Medicine Affiliated Hospital (Institutional Review Board No. 2022KL-051) and Sichuan Cancer Hospital (Institutional Review Board No. SCCHEC-02–2022-109). Written informed consent was obtained from all patients prior to the surgery. Animal experiments were approved by the Institutional Animal Care and Use Committees of Hospital of Chengdu University of Traditional Chinese Medicine. Consent for publication: We have obtained consents to publish this paper from all the participants of this study. Competing interests: The authors declare no competing interests.
Figures





References
-
- Mazzone PJ, Lam L. Evaluating the Patient With a Pulmonary Nodule: A Review. JAMA. 2022;327(3):264–73. - PubMed
-
- Ye T, Deng L, Wang S, Xiang J, Zhang Y, Hu H, et al. Lung Adenocarcinomas Manifesting as Radiological Part-Solid Nodules Define a Special Clinical Subtype. J Thorac Oncol. 2019;14(4):617–27. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

