High CD44 expression and enhanced E-selectin binding identified as biomarkers of chemoresistant leukemic cells in human T-ALL
- PMID: 39580584
- PMCID: PMC11794132
- DOI: 10.1038/s41375-024-02473-7
High CD44 expression and enhanced E-selectin binding identified as biomarkers of chemoresistant leukemic cells in human T-ALL
Abstract
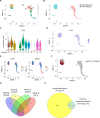
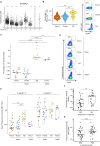
T-cell acute lymphoblastic leukemia (T-ALL) is a hematopoietic malignancy characterized by increased proliferation and incomplete maturation of T-cell progenitors, for which relapse is often of poor prognosis. To improve patient outcomes, it is critical to understand the chemoresistance mechanisms arising from cell plasticity induced by the bone marrow (BM) microenvironment. Single-cell RNA sequencing of human T-ALL cells from adipocyte-rich and adipocyte-poor BM revealed a distinct leukemic cell population defined by quiescence and high CD44 expression (Ki67neg/lowCD44high). During in vivo treatment, these cells evaded chemotherapy, and were further called Chemotherapy-resistant Leukemic Cells (CLCs). Patient sample analysis revealed Ki67neg/lowCD44high CLCs at diagnosis and during relapse, with each displaying a specific transcriptomic signature. Interestingly, CD44high expression in T-ALL Ki67neg/low CLCs was associated with E-selectin binding. Analysis of 39 human T-ALL samples revealed significantly enhanced E-selectin binding activity in relapse/refractory samples compared with drug-sensitive samples. These characteristics of chemoresistant T-ALL CLCs provide key insights for prognostic stratification and novel therapeutic options.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16:494–507. - PubMed
-
- Vadillo E, Dorantes-Acosta E, Pelayo R, Schnoor M. T cell acute lymphoblastic leukemia (T-ALL): New insights into the cellular origins and infiltration mechanisms common and unique among hematologic malignancies. Blood Rev. 2018;32:36–51. - PubMed
-
- Schelker RC, Iberl S, Müller G, Hart C, Herr W, Grassinger J. TGF-β1 and CXCL12 modulate proliferation and chemotherapy sensitivity of acute myeloid leukemia cells co-cultured with multipotent mesenchymal stromal cells. Hematology. 2018;23:337–45. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

