Role of m6A Methylation Regulators in the Diagnosis and Subtype Classification of COPD Based on the GEO Database
- PMID: 39580709
- PMCID: PMC11585962
- DOI: 10.1111/jcmm.70226
Role of m6A Methylation Regulators in the Diagnosis and Subtype Classification of COPD Based on the GEO Database
Abstract
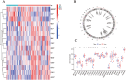
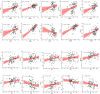
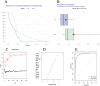
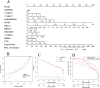
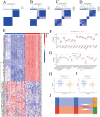
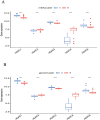
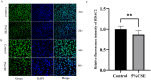
N6-methyladenosine (m6A) is a prevalent mRNA modifier, yet its role in chronic obstructive pulmonary disease (COPD) remains unexplored. We sourced expression levels of m6A methylation regulators from the GSE76925 dataset. These regulators' differential expression (DEMs) predicted COPD risk via random forest and support vector machine models. Additionally, a nomogram model using DEMs estimated COPD prevalence. We employed consistent cluster analysis of m6A methylation regulators to categorise COPD samples into distinct subtypes. Analyses of immune cell infiltration in these subtypes and differential gene expression (DEGs) across m6A methylation subtypes were conducted. A cell model validated several m6A regulators and their associated pathways. Fifteen m6A methylation regulators showed differential expression and were used in random forest and support vector machine models. Eleven were selected for a nomogram model, which decision curve analysis suggested could benefit patients. Consensus cluster analysis divided the COPD samples into two subtypes: Cluster A and Cluster B. Cluster B was associated with neutrophil and eosinophil-dominated immunity, while Cluster A was linked with monocyte-dominated immunity. Validation of some research findings was achieved through cell experiments. m6A methylation regulators appear instrumental in diagnosing and classifying subtypes of COPD.
Keywords: COPD; diagnosis; differential genes; m6A methylation regulators; subtype.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Wang Z., Locantore N., Haldar K., et al., “Inflammatory Endotype–Associated Airway Microbiome in Chronic Obstructive Pulmonary Disease Clinical Stability and Exacerbations: A Multicohort Longitudinal Analysis,” American Journal of Respiratory and Critical Care Medicine 203 (2021): 1488–1502, 10.1164/rccm.202009-3448OC. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

