High-affinity agonists reveal recognition motifs for the MRGPRD GPCR
- PMID: 39580805
- PMCID: PMC12006980
- DOI: 10.1016/j.celrep.2024.114942
High-affinity agonists reveal recognition motifs for the MRGPRD GPCR
Abstract
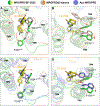
The human MRGPRD protein is a member of the Mas-related G protein-coupled receptors (MRGPRs) that is involved in the sensing of pain, itch, and other inflammatory stimuli. As with other MRGPRs, MRGPRD is a relatively understudied receptor with few known agonists. The most potent small-molecule agonist of MRGPRD reported so far is β-alanine, with an affinity in the micromole range, which largely restricts its functional study. Here, we report two MRGPRD agonists, EP-2825 and EP-3945, that are approximately 100-fold more potent than β-alanine and determine the structures of MRGPRD-Gq in complex with EP-2825 and EP-3945, respectively. The structures reveal distinct agonist binding modes of MRGPRD and large conformational plasticity of the orthosteric pocket. Collectively, the discovery of high-affinity MRGPRD agonists and their distinct binding modes will facilitate the functional study and the structure-based design of ligands targeting this understudied receptor.
Keywords: CP: Molecular biology; GPCR; MRGPRD; agonist recognition; drug discovery; structure.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A pending patent application has been filed by Escient Pharmaceuticals that includes compounds EP-2825 and EP-3945. B.L.R. is on the scientific advisory board of Escient Pharmaceuticals.
Figures
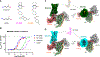



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

