Transcriptomic data reveals MYC as an upstream regulator in laying hen follicular recruitment
- PMID: 39580902
- PMCID: PMC11625332
- DOI: 10.1016/j.psj.2024.104547
Transcriptomic data reveals MYC as an upstream regulator in laying hen follicular recruitment
Abstract
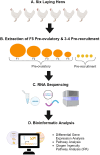
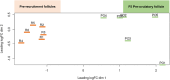
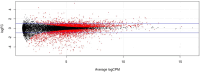
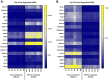
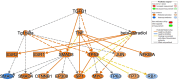
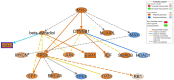
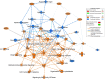
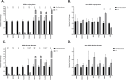
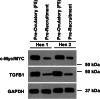
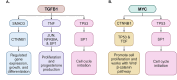
Understanding the mechanisms of follicular recruitment is essential for improving laying hen and broiler breeder productivity, as it directly influences egg production. Despite advancements in poultry breeding for enhanced egg production, the factors driving successful ovarian follicle maturation remain inadequately understood. This study investigates the genetic drivers mediating the transition of pre-recruitment follicles to the pre-ovulatory phase, a crucial stage before ovulation. Using RNA sequencing and bioinformatics approaches such as a differential gene expression analysis, we compared pre-recruitment follicles with the recently recruited F5 pre-ovulatory follicle to identify key genes and upstream regulators involved in this transition. Further validation through qRT-PCR confirmed these findings. Using Qiagen's Ingenuity Pathway Analysis we identified MYC proto-oncogene (C-Myc) as a pivotal upstream regulator, controlling genes essential for cell proliferation and differentiation. Additionally, TGFβ1 emerged as a key regulator, influencing pathways involving SMAD3, TNF, and TP53. The study highlights the intricate regulatory network involving MYC and other transcription factors such as CTNNB1, crucial for follicular development. These findings provide valuable insights into the molecular mechanisms governing follicular selection and maturation, which are essential for enhancing egg production efficiency. Future research should explore the roles of MYC, CTNNB1, and other driver genes in follicular development to further understand and improve reproductive efficiency in poultry.
Keywords: Broiler breeder; Follicular recruitment; Layer hen; MYC; Ovary.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Kathryn M Ellwood reports financial support was provided by USDA. Aditya Dutta reports financial support was provided by National Institutes of Health. Aditya Dutta reports financial support was provided by University of Delaware Research Foundation Inc. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Cross-species regulatory network analysis identifies FOXO1 as a driver of ovarian follicular recruitment.Sci Rep. 2024 Dec 28;14(1):30787. doi: 10.1038/s41598-024-80003-2. Sci Rep. 2024. PMID: 39730395 Free PMC article.
-
Transcriptomic analysis of ovarian follicles uncovers the crucial genes relevant to follicle selection and preovulatory hierarchy in hens.J Anim Sci. 2023 Jan 3;101:skad241. doi: 10.1093/jas/skad241. J Anim Sci. 2023. PMID: 37453139 Free PMC article.
-
Transcriptome comparative analysis of ovarian follicles reveals the key genes and signaling pathways implicated in hen egg production.BMC Genomics. 2021 Dec 15;22(1):899. doi: 10.1186/s12864-021-08213-w. BMC Genomics. 2021. PMID: 34911438 Free PMC article.
-
The domestic chicken: Causes and consequences of an egg a day.Poult Sci. 2015 Apr;94(4):816-20. doi: 10.3382/ps/peu083. Epub 2015 Feb 9. Poult Sci. 2015. PMID: 25667424 Review.
-
Chicken ovarian follicular atresia: interaction network at organic, cellular, and molecular levels.Poult Sci. 2024 Aug;103(8):103893. doi: 10.1016/j.psj.2024.103893. Epub 2024 May 26. Poult Sci. 2024. PMID: 38870615 Free PMC article. Review.
Cited by
-
Identifying candidate genetic variants for egg number by analyzing over 1,000 fully sequenced layers.Gigascience. 2025 Jan 6;14:giaf064. doi: 10.1093/gigascience/giaf064. Gigascience. 2025. PMID: 40526492 Free PMC article.
-
Cross-species regulatory network analysis identifies FOXO1 as a driver of ovarian follicular recruitment.Sci Rep. 2024 Dec 28;14(1):30787. doi: 10.1038/s41598-024-80003-2. Sci Rep. 2024. PMID: 39730395 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

