BioPAX in 2024: Where we are and where we are heading
- PMID: 39582893
- PMCID: PMC11585474
- DOI: 10.1016/j.csbj.2024.10.045
BioPAX in 2024: Where we are and where we are heading
Abstract
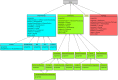
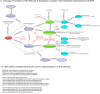
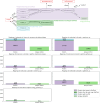
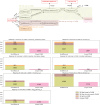
In systems biology, the study of biological pathways plays a central role in understanding the complexity of biological systems. The massification of pathway data made available by numerous online databases in recent years has given rise to an important need for standardization of this data. The BioPAX format (Biological Pathway Exchange) emerged in 2010 as a solution for standardizing and exchanging pathway data across databases. BioPAX is a Semantic Web format associated to an ontology. It is highly expressive, allowing to finely describe biological pathways at the molecular and cellular levels, but the associated intrinsic complexity may be an obstacle to its widespread adoption. Here, we report on the use of the BioPAX format in 2024. We compare how the different pathway databases use BioPAX to standardize their data and point out possible avenues for improvement to make full use of its potential. We also report on the various tools and software that have been developed to work with BioPAX data. Finally, we present a new concept of abstraction on BioPAX graphs that would allow to specifically target areas in a BioPAX graph needed for a specific analysis, thus differentiating the format suited for representation and the abstraction suited for contextual analysis.
Keywords: BioPAX; Biological pathways; Databases; Systems Biology.
© 2024 The Authors.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures




Similar articles
-
BioPAX support in CellDesigner.Bioinformatics. 2011 Dec 15;27(24):3437-8. doi: 10.1093/bioinformatics/btr586. Epub 2011 Oct 21. Bioinformatics. 2011. PMID: 22021903 Free PMC article.
-
BioPAX-Parser: parsing and enrichment analysis of BioPAX pathways.Bioinformatics. 2020 Aug 1;36(15):4377-4378. doi: 10.1093/bioinformatics/btaa529. Bioinformatics. 2020. PMID: 32437515
-
Kinetic Modeling using BioPAX ontology.Proceedings (IEEE Int Conf Bioinformatics Biomed). 2007 Nov 2;2007:339-348. doi: 10.1109/BIBM.2007.55. Proceedings (IEEE Int Conf Bioinformatics Biomed). 2007. PMID: 20862270 Free PMC article.
-
e-Science and biological pathway semantics.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S3. doi: 10.1186/1471-2105-8-S3-S3. BMC Bioinformatics. 2007. PMID: 17493286 Free PMC article. Review.
-
PAX of mind for pathway researchers.Drug Discov Today. 2005 Jul 1;10(13):937-42. doi: 10.1016/S1359-6446(05)03501-4. Drug Discov Today. 2005. PMID: 15993813 Review.
References
Publication types
LinkOut - more resources
Full Text Sources
