Single-cell data revealed the function of natural killer cells and macrophage cells in chemotherapy tolerance in acute myeloid leukemia
- PMID: 39583114
- PMCID: PMC11586048
- DOI: 10.7717/peerj.18521
Single-cell data revealed the function of natural killer cells and macrophage cells in chemotherapy tolerance in acute myeloid leukemia
Abstract
Background: Acute myeloid leukemia (AML) is highly prevalent and heterogeneous among adult acute leukemias. Current chemotherapeutic approaches for AML often face the challenge of drug resistance, and AML immune cells play an important role in the regulation of AML drug resistance. Thus, it is of key significance to explore the regulatory mechanisms of immune cells in AML to alleviate chemotherapy resistance in AML.
Methods: Based on AML single-cell transcriptomic data, this study revealed the differences in the expression of immune cell subpopulations and marker genes in AML patients in the complete remission group (CR) compared to AML patients in the non-complete remission group (non-CR) after chemotherapy. Functional enrichment by clusterprofiler revealed the regulatory functions of differentially expressed genes (DEGs) in AML. AUCell enrichment scores were used to assess the immunoregulatory functions of immune cells. Pseudotime analysis was used to construct immune cell differentiation trajectories. CellChat was used for cellular communication analysis to elucidate the interactions between immune cells. Survival analysis with the R package "survival" revealed the role of immune cell marker genes on AML prognosis. Finally, the wound healing and trans-well assay were performed.
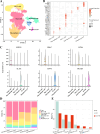
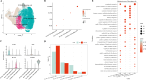
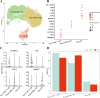
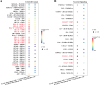
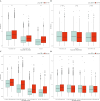
Results: Single-cell clustering analysis revealed that NK/T cells and macrophage cells subpopulations were significantly higher in non-CR AML patients than in CR AML. AUCell enrichment analysis revealed that FCAR+ and FCGR3A+ macrophages were significantly more active in the non-CR group and correlated with processes regulating cellular energy metabolism and immune cell activity. Differentially expressed NK cell marker genes between CR and non-CR groups mainly included HBA1, S100A8, and S100A9, which were associated with cancer drug resistance regulation, these marker genes of (FCAR, FCGR3A, PREX1, S100A8 and S100A9) were upregulated in human chronic myeloid leukemia cells (HAP1) and silencing of S100A8 affected migration and invasion of HAP1 cells. In particular, the differentiation pathways of macrophages and NK cells in non-CR differed from those of patients in the CR group. Cellular communication analyses showed that ligand-receptor pairs between NK cells and macrophage cells mainly included HLA-E-KLRK1, HLA-E-KLRC1, HLA-E-CD94:NKG2A, CLEC2B-KLRB1. In addition, LGALS9-CD45, CCL3L1- CCR1, CCL3-CCR1 between these two immune cells mainly regulate secreted signaling to mediate AML progression. Marker genes in NK/T cells and macrophage cells were significantly associated with AML prognosis.
Conclusion: This study reveals the potential role of NK cells and macrophages in AML chemoresistance through the analysis of single-cell RNA sequencing data. This provides new ideas and insights into the key mechanisms of immune cells in AML treatment.
Keywords: Acute myeloid leukemia; Chemotherapy resistance; Macrophage cells; Natural killer cells; Single cell RNA-seq.
©2024 Gao et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



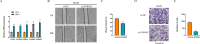
References
-
- Bottcher M, Panagiotidis K, Bruns H, Stumpf M, Volkl S, Geyh S, Dietel B, Schroeder T, Mackensen A, Mougiakakos D. Bone marrow stroma cells promote induction of a chemoresistant and prognostic unfavorable S100A8/A9high AML cell subset. Blood Advances. 2022;6(21):5685–5697. doi: 10.1182/bloodadvances.2021005938. - DOI - PMC - PubMed
-
- Bruchard M, Mignot G, Derangere V, Chalmin F, Chevriaux A, Vegran F, Boireau W, Simon B, Ryffel B, Connat JL, Kanellopoulos J, Martin F, Rebe C, Apetoh L, Ghiringhelli F. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nature Medicine. 2013;19(1):57–64. doi: 10.1038/nm.2999. - DOI - PubMed
-
- Chen Z, Zhao M, Li M, Sui Q, Bian Y, Liang J, Hu Z, Zheng Y, Lu T, Huang Y, Zhan C, Jiang W, Wang Q, Tan L. Identification of differentially expressed genes in lung adenocarcinoma cells using single-cell RNA sequencing not detected using traditional RNA sequencing and microarray. Laboratory Investigation; a Journal of Technical Methods and Pathology. 2020;100(10):1318–1329. doi: 10.1038/s41374-020-0428-1. - DOI - PubMed
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

