Association between genetic clades and cancer prevalence suggested by French-wide study of oncogenic small ruminant β-retrovirus diversity
- PMID: 39583158
- PMCID: PMC11582038
- DOI: 10.3389/fcimb.2024.1466333
Association between genetic clades and cancer prevalence suggested by French-wide study of oncogenic small ruminant β-retrovirus diversity
Abstract
Introduction: ENTV (Enzootic Nasal Tumor Virus) and JSRV (Jaagsiekte Sheep Retrovirus) are β-retroviruses responsible for respiratory cancers in sheep and goats. In this study, we analyzed the genetic features of the sheep and goat β-Retroviruses (29 JSRV and 24 ENTV strains) circulating in France to identify molecular signatures associated with disease severity in flocks.
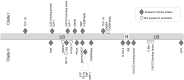
Methods: We developed a highly specific PCR to amplify and sequence exogenous targeted regions or near full length proviruses based on limited discriminating motifs along their genomes.
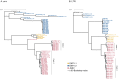
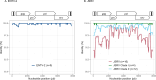
Results: The phylogenetic reconstructions based on the Long Terminal Repeat (LTR) and env regions suggest that one major strain is circulating on the French territory for ENTV-1 and ENTV-2 while not clustering with already published Spanish, Canadian or Chinese strains. JSRV strains circulating in French sheep flocks were distributed in 2 distinct genetic clades clustering with sequences originating from North America, Africa and United-Kingdom. JSRV clade I was found to be associated with a higher incidence of cancer in French flocks. Specific motifs spanning the entire JSRV genome particularly in the LTRs and in the intracytoplasmic domain of the envelope were detected between the two genetic subtypes.
Discussion: This work represents the first nationwide study describing the circulation of the three closely related β-oncogenic retroviruses JSRV, ENTV-1 and ENTV-2 in French sheep and goat flocks. Better characterization of strain genetics is a critical step in monitoring circulating - retroviruses, especially those associated with higher cancer incidence in small ruminants.
Keywords: ENTV-1; ENTV-2; JSRV; genomic epidemiology; virulence factors.
Copyright © 2024 Riocreux-Verney, Verneret, Diesler, Dolmazon, Gineys, Cadoré, Turpin and Leroux.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- BBMap (2023). SourceForge. Available online at: https://sourceforge.net/projects/bbmap/ (Accessed March 26, 2024).
-
- Caporale M., Centorame P., Giovannini A., Sacchini F., Di Ventura M., de las Heras M., et al. (2005). Infection of lung epithelial cells and induction of pulmonary adenocarcinoma is not the most common outcome of naturally occurring JSRV infection during the commercial lifespan of sheep. Virology 338, 144–153. doi: 10.1016/j.virol.2005.05.018 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

