A simple MiMIC-based approach for tagging endogenous genes to visualise live transcription in Drosophila
- PMID: 39584418
- PMCID: PMC11701513
- DOI: 10.1242/dev.204294
A simple MiMIC-based approach for tagging endogenous genes to visualise live transcription in Drosophila
Abstract
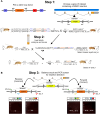
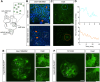
Live imaging of transcription in the Drosophila embryo using the MS2 or PP7 systems is transforming our understanding of transcriptional regulation. However, insertion of MS2/PP7 stem-loops into endogenous genes requires laborious CRISPR genome editing. Here, we exploit the previously described Minos-mediated integration cassette (MiMIC) transposon system in Drosophila to establish a method for simply and rapidly inserting MS2/PP7 cassettes into any of the thousands of genes carrying a MiMIC insertion. In addition to generating a variety of stem-loop donor fly stocks, we have made new stocks expressing the complementary coat proteins fused to different fluorescent proteins. We show the utility of this MiMIC-based approach by MS2/PP7 tagging of endogenous genes and the long non-coding RNA roX1, then imaging their transcription in living embryos. We also present live transcription data from larval brains, the wing disc and ovary, thereby extending the tissues that can be studied using the MS2/PP7 system. Overall, this first high-throughput method for tagging mRNAs in Drosophila will facilitate the study of transcription dynamics of thousands of endogenous genes in a range of Drosophila tissues.
Keywords: Drosophila; Live imaging; MS2-MCP system; MiMIC; PP7-PCP system; Transcription.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Bellen, H. J., Levis, R. W., Liao, G., He, Y., Carlson, J. W., Tsang, G., Evans-Holm, M., Hiesinger, P. R., Schulze, K. L., Rubin, G. M.et al. (2004). The BDGP Gene Disruption Project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761-781. 10.1534/genetics.104.026427 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

