Differential Effects of Extracellular Vesicles from Two Different Glioblastomas on Normal Human Brain Cells
- PMID: 39585062
- PMCID: PMC11587087
- DOI: 10.3390/neurolint16060103
Differential Effects of Extracellular Vesicles from Two Different Glioblastomas on Normal Human Brain Cells
Abstract
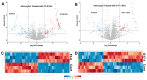
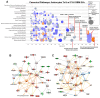
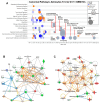
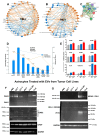
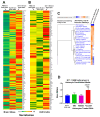
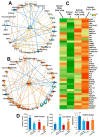
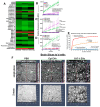
Background/Objectives: Glioblastomas (GBMs) are dreadful brain tumors with abysmal survival outcomes. GBM extracellular vesicles (EVs) dramatically affect normal brain cells (largely astrocytes) constituting the tumor microenvironment (TME). We asked if EVs from different GBM patient-derived spheroid lines would differentially alter recipient brain cell phenotypes. This turned out to be the case, with the net outcome of treatment with GBM EVs nonetheless converging on increased tumorigenicity. Methods: GBM spheroids and brain slices were derived from neurosurgical patient tissues following informed consent. Astrocytes were commercially obtained. EVs were isolated from conditioned culture media by ultrafiltration, concentration, and ultracentrifugation. EVs were characterized by nanoparticle tracking analysis, electron microscopy, biochemical markers, and proteomics. Astrocytes/brain tissues were treated with GBM EVs before downstream analyses. Results: EVs from different GBMs induced brain cells to alter secretomes with pro-inflammatory or TME-modifying (proteolytic) effects. Astrocyte responses ranged from anti-viral gene/protein expression and cytokine release to altered extracellular signal-regulated protein kinase (ERK1/2) signaling pathways, and conditioned media from EV-treated cells increased GBM cell proliferation. Conclusions: Astrocytes/brain slices treated with different GBM EVs underwent non-identical changes in various omics readouts and other assays, indicating "personalized" tumor-specific GBM EV effects on the TME. This raises concern regarding reliance on "model" systems as a sole basis for translational direction. Nonetheless, net downstream impacts from differential cellular and TME effects still led to increased tumorigenic capacities for the different GBMs.
Keywords: astrocytes; extracellular vesicles; glioblastoma; innate immunity; protease; proteomics; signaling; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- Wirsching H.G., Galanis E., Weller M. Glioblastoma. Handb. Clin. Neurol. 2016;134:381–397. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

