Genetic diversity and virulence variability of Sclerotinia sclerotiorum in Eastern and Northeastern India
- PMID: 39585888
- PMCID: PMC11588274
- DOI: 10.1371/journal.pone.0312472
Genetic diversity and virulence variability of Sclerotinia sclerotiorum in Eastern and Northeastern India
Abstract
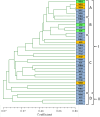
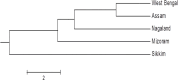
Sclerotinia sclerotiorum, the necrotrophic cosmopolitan fungus, has become an emerging and re-emerging pathogen in the subtropical regions. Genetic diversity of 36 isolates of the fungus isolated from infected samples collected from the eastern and North eastern states was carried out using UP-PCR and SSR. Virulence variability was analysed based on four different measures. Among the eight UP-PCR primers and various combinations used, L-21, 3-2 and AA2M2-AS4 generated maximum number of fingerprints (13, 13 and 12, respectively) ranging from 100bp to 1kb. The isolates exhibited varied level of aggressiveness; majority (77.78%) were moderately virulent, 8.33% (22.22% of Assam and 6.67% of West Bengal) isolates were highly virulent, and 13.89% were less virulent. Several amplification products viz., 500bp generated by AA2M2-AS4, 150bp by AA2M2-L-21 and 100bp by L-21-3-2 were positively correlated with disease severity grading at 5% level of significance, whereas, 600bp band generated by AA2M2-3-2 was correlated at 1% level of significance. This indicates presence of these bands in highly virulent isolates. Out of the eight SSR primers, TATG9 did not generate any amplification and the isolates were divided into two major groups; the group II contained single isolate from Nagaland (NG4) indicating it to be genetically diverse from rest of the isolates. The subgroup A of the major group I was the largest and most diverse group with 11 members indicating genetic admixture within different geographic populations with different levels of similarity (70-100%). Genetic diversity based on SSR banding pattern showed highest value of Nei's gene diversity and Shannon's index of diversity (%pb = 61.11; h = 0.219; I = 0.330) for the Nagaland population with 9 members followed by West Bengal population with 15 members. Nei's genetic distance of all the tested populations was low, ranging from 0.0014 to 0.2350; however, genetic identity was high ranging from 0.7905 to 0.9986. The findings suggest that the pathogen populations of eastern and North eastern region were predominantly clonal with some evidence of infrequent out crossing.
Copyright: © 2024 Borah et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Purdy LH (1979) Sclerotinia sclerotium: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology69: 875–880.
-
- Bolton MD, Thomma BPHJ, Nelson BD, (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Molecular Plant Pathology 7:1–16 - PubMed
-
- Saharan GS, Mehta N, (2008) Sclerotinia diseases of crop plants: Biology, ecology and disease management. Springer Netherlands: 481.
-
- Dutta S, Borah Tasvina R, Barman AR, Hansda S, Ghosh PP, (2016) Overview of Sclerotinia sclerotiorum in India with special reference to its emerging severity in Eastern Region. Indian Phytopathlogy 69(4s): 260–265.
-
- Borah TR, Dutta S, Barman AR, Helim R, Sen K, (2021) Variability and host range of Sclerotinia sclerotiorumin eastern and North eastern India. Journal of Plant Pathology 103: 809–822. doi: 10.1007/s42161-021-00815-3 - DOI
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

